Engineering Biosynthesis: Synthetic Biology Approaches for Sustainable Magnolol Production
This article comprehensively reviews the application of synthetic biology to address critical challenges in magnolol production.
Engineering Biosynthesis: Synthetic Biology Approaches for Sustainable Magnolol Production
Abstract
This article comprehensively reviews the application of synthetic biology to address critical challenges in magnolol production. It explores the foundational biosynthesis pathway of magnolol in Magnolia officinalis, detailing the recent identification of the pivotal laccase enzyme MoLAC14. The content covers methodological advances in engineering microbial cell factories and optimizing protein expression for biosynthesis. It further discusses strategies for troubleshooting production bottlenecks, such as enhancing enzyme thermostability and activity through rational design. Finally, the article provides a comparative analysis of synthetic biology-derived magnolol against traditional extraction and chemical synthesis, validating its potential to provide a scalable, sustainable, and high-purity supply of this versatile therapeutic compound for biomedical research and drug development.
Decoding Nature's Blueprint: The Biosynthetic Pathway and Therapeutic Promise of Magnolol
Magnolol (MG), a hydroxylated biphenyl compound derived from the bark of Magnolia officinalis, has garnered significant interest for its broad-spectrum pharmacological activities, including anti-cancer, anti-inflammatory, and neuroprotective effects [1] [2] [3]. As a natural polyphenol, magnolol exhibits a unique pharmacophore structure consisting of two hydroxylated aromatic rings connected by a single C–C bond, enabling interactions with numerous proteins and contributing to its diverse biological activity [3]. However, its clinical translation faces challenges, primarily due to poor water solubility and low bioavailability [3] [4]. Synthetic biology emerges as a promising approach to overcome these limitations by enabling efficient and sustainable production of magnolol and its semi-synthetic derivatives [5]. This application note delineates the mechanistic underpinnings of magnolol's pharmacological actions, provides detailed experimental protocols for evaluating its efficacy, and explores its therapeutic potential within the context of advanced production platforms.
Anti-Cancer Mechanisms and Quantitative Profiling
Magnolol demonstrates efficacy against a diverse range of cancers by targeting multiple hallmarks of cancer progression. The compound exerts its effects through modulation of critical signaling pathways, induction of programmed cell death, and disruption of metabolic processes.
Table 1: Anti-Cancer Mechanisms of Magnolol Across Various Cancer Types
| Cancer Type | Molecular Targets & Mechanisms | Experimental Models | Key Quantitative Findings |
|---|---|---|---|
| Oral Squamous Cell Carcinoma (OSCC) | Inhibits EGFR/NF-κB axis; induces caspase-3, -8, -9 cleavage; promotes M1 macrophages & dendritic cells [6]. | MOC1-bearing animal models [6]. | Tumor growth suppression; increased activated cytotoxic T cells; reduced phosphorylation of EGFR/NF-κB [6]. |
| Multiple Cancers (e.g., Bladder, Colon, Liver, Lung) | Modulates NF-κB, MAPK, PI3K/Akt/mTOR pathways; induces apoptosis & autophagy; inhibits DNA synthesis [1] [3]. | Various cell lines & animal models [1] [3]. | Effective against numerous cancer types; induces apoptosis in liver & colon cancer cells; promotes autophagy in lung cancer cells [1] [3]. |
| Gallbladder Cancer | Suppresses cancer cell growth via p53 pathway [3]. | Cell line studies [3]. | Growth inhibition via p53-mediated mechanisms [3]. |
| Via Mitochondrial Targeting | Disrupts mitochondrial electron transport chain (ETC); increases ROS; induces mitophagy & apoptosis [7] [4]. | C. elegans; A. suum larvae; various tumor cells [7] [4]. | EC₅₀ vs. A. suum: 11.08 μM [7]; Derivative MTP showed enhanced cytotoxicity vs. parent MAG [4]. |
The following diagram illustrates the core anti-cancer signaling pathways modulated by magnolol and its derivatives:
Synthetic Biology and Biosynthesis Protocols
The sustainable production of magnolol is critical for ongoing research and therapeutic development. Traditional extraction from Magnolia officinalis bark is constrained by low yields (approximately 1-10%) and a long cultivation cycle of 10-15 years [5] [3]. Chemical synthesis often suffers from low yield, lack of specificity, and environmental concerns [5]. Synthetic biology offers a viable alternative.
Proposed Biosynthetic Pathway and Key Enzyme Identification
Research has proposed a biosynthetic pathway where magnolol is synthesized from chavicol, a precursor derived from tyrosine. The conversion is catalyzed by a key enzyme, laccase (MoLAC14) [5].
Table 2: Research Reagent Solutions for Magnolol Biosynthesis and Analysis
| Reagent / Tool | Function / Application | Specifications / Notes |
|---|---|---|
| MoLAC14 (Laccase Enzyme) | Catalyzes the oxidative coupling of two chavicol molecules into magnolol [5]. | Key identified enzyme from M. officinalis; can be engineered for improved stability (e.g., E345P, G377P, L532A mutations) [5]. |
| pET-28a Vector | Expression vector for heterologous production of laccase enzymes in microbial systems [5]. | Used for cloning and expressing MoLAC14 and other candidate laccase genes [5]. |
| Chavicol | Direct precursor substrate for magnolol synthesis in the proposed enzymatic pathway [5]. | Proposed to be synthesized from tyrosine via enzymes like TAL, 4CL, CCR, and ADH [5]. |
| HPLC with PDA Detector | Analysis and quantification of magnolol produced in enzymatic reactions or extracted from samples [5] [7]. | Used for high-resolution antiparasitic profiling and validating magnolol synthesis [7]. |
| S. cerevisiae or E. coli | Model host organisms for the heterologous production of magnolol using synthetic biology approaches [5]. | Engineered microbial chassis for biosynthetic production. |
Protocol: In Vitro Enzymatic Synthesis of Magnolol
This protocol outlines the key steps for producing magnolol using the identified laccase enzyme, MoLAC14 [5].
- Objective: To catalyze the conversion of chavicol into magnolol using recombinantly expressed MoLAC14 laccase.
Materials:
- Cloned Vector: pET-28a vector containing the codon-optimized MoLAC14 gene (or mutant variants like L532A for higher yield).
- Expression Host: E. coli BL21(DE3) competent cells.
- Culture Media: LB broth supplemented with appropriate antibiotic (e.g., Kanamycin, 50 µg/mL).
- Inducer: Isopropyl β-d-1-thiogalactopyranoside (IPTG).
- Substrate: Chavicol.
- Buffers: Lysis buffer (e.g., PBS or Tris-HCl with lysozyme), purification buffers (if performing protein purification via His-tag).
- Analysis Equipment: HPLC system with a C18 column and PDA detector.
Procedure:
- Heterologous Expression: Transform the pET-28a-MoLAC14 plasmid into E. coli BL21(DE3). Grow the culture in LB medium at 37°C until OD₆₀₀ reaches ~0.6. Induce protein expression by adding IPTG to a final concentration of 0.1-0.5 mM and incubate further at a lower temperature (e.g., 16-18°C) for 16-20 hours.
- Enzyme Preparation: Harvest cells by centrifugation. Lyse the cell pellet using sonication or lysis buffer. The crude lysate containing the enzyme can be used directly, or the enzyme can be purified using affinity chromatography (if a His-tag is present).
- Enzymatic Reaction: Set up a reaction mixture containing:
- Buffer (e.g., phosphate buffer, 50-100 mM, pH ~7.0)
- Chavicol substrate (concentration to be optimized, e.g., 1-10 mM)
- Enzyme preparation (crude lysate or purified protein)
- Incubate at 30-37°C with shaking for several hours.
- Reaction Termination & Analysis: Stop the reaction by adding an equal volume of methanol or acetonitrile. Remove precipitates by centrifugation. Analyze the supernatant using HPLC.
- HPLC Conditions (example): C18 column; mobile phase: gradient of water (with 0.1% formic acid) and acetonitrile (with 0.1% formic acid); flow rate: 0.5-1.0 mL/min; detection: UV at 254-290 nm. Compare the retention time and spectrum with a magnolol standard.
- Validation: Confirm magnolol identity using Mass Spectrometry (MS) by comparing the mass-to-charge ratio with the standard.
Notes: The mutant MoLAC14 (L532A) has been reported to significantly enhance magnolol production, reaching levels up to 148.83 mg/L in vitro [5].
The workflow for magnolol production and validation, from gene identification to final analysis, is depicted below:
Experimental Protocols for Therapeutic Efficacy Evaluation
Protocol: Evaluating Magnolol as a Radiosensitizer in Oral Cancer Models
This protocol describes the in vivo evaluation of magnolol's efficacy in enhancing radiotherapy for oral squamous cell carcinoma (OSCC) [6].
- Objective: To assess the combined effect of magnolol and radiation therapy (RT) on tumor growth, apoptosis, and immune modulation in a murine OSCC model.
Materials:
- Animals: 6-8 week-old male C57BL/6 mice.
- Cell Line: MOC1 OSCC cells.
- Reagents: Magnolol (for gavage), Erlotinib (positive control), DMSO, Matrigel, Isoflurane/anesthetic cocktail (Zoletil 50/Xylazine).
- Equipment: Linear accelerator for radiotherapy, Flow cytometer, Equipment for immunohistochemistry (IHC).
- Antibodies: For flow cytometry (e.g., CD8, IFN-γ, IL-2, CD11b, CD11c, Ly-6G/Ly-6C) and IHC (e.g., Cleaved Caspase-3, -8, -9) [6].
Procedure:
- Tumor Inoculation: Harvest and resuspend MOC1 cells in a 3:7 mixture of culture medium and Matrigel. Subcutaneously inject 1 x 10ⶠcells in a 100 µL volume into the right cheek of each anesthetized mouse.
- Group Allocation and Treatment: When the average tumor volume reaches ~60 mm³, randomize mice into five groups (n=5/group):
- Group 1 (Control): Daily gavage with vehicle (0.1% DMSO in water).
- Group 2 (Magnolol alone): Daily gavage with magnolol (40 mg/kg/day).
- Group 3 (RT alone): Single dose of 6 Gy radiation to the tumor site on day 1.
- Group 4 (Magnolol + RT): Daily gavage with magnolol (40 mg/kg/day) starting on day 0, plus a single dose of 6 Gy on day 1.
- Group 5 (Erlotinib + RT): Daily gavage with erlotinib (20 mg/kg/day) starting on day 0, plus a single dose of 6 Gy on day 1.
- Monitoring: Measure tumor dimensions bi-daily using callipers. Calculate volume as (Height × Width² × 0.523). Monitor animal body weight as an indicator of general health and toxicity.
- Terminal Analysis: On day 24, euthanize mice and collect samples:
- Tumors: Weigh and record. Divide for IHC (fixed in formalin) and flow cytometry (single-cell suspension for immune profiling).
- Spleen and Lymph Nodes: Process into single-cell suspensions for flow cytometry.
- Immune Profiling by Flow Cytometry: Stain cells with specific antibody panels.
- Cytotoxic T cells: Surface stain for CD8, intracellular stain for IFN-γ and IL-2 after stimulation.
- Macrophages/Dendritic Cells: Surface stain for M1 (e.g., CD11c, CD86) and M2 (e.g., CD206) markers.
- Myeloid-Derived Suppressor Cells (MDSCs) & Tregs: Stain for CD11b, Ly-6G/Ly-6C, and FoxP3.
- Immunohistochemistry (IHC): Perform IHC on tumor sections for cleaved caspase-3, -8, and -9 to assess apoptosis, and for phospho-EGFR and phospho-NF-κB to evaluate pathway inhibition.
Key Outcome Measures:
- Tumor growth curves and final tumor weight.
- Flow cytometry data on immune cell populations within the tumor microenvironment (TME).
- IHC scoring for apoptosis markers and signaling pathway activation.
Protocol: Antiparasitic Screening Assay for Magnolol
This protocol details an in vitro assay to evaluate the anthelmintic (anti-parasitic worm) efficacy of magnolol [7].
- Objective: To determine the efficacy (ECâ‚…â‚€) of magnolol against the larval stage of the porcine roundworm Ascaris suum, a model parasite.
Materials:
- Parasite Material: Fresh Ascaris suum adults from a slaughterhouse. Embryonated eggs are isolated and hatched to obtain larvae.
- Reagents: Magnolol (dissolved in DMSO), Ivermectin (positive control), Hanks' Balanced Salt Solution (HBSS), RPMI-1640 Medium, Penicillin-Streptomycin, Amphotericin B.
- Equipment: COâ‚‚ incubator, Sonicated water bath, Baermann apparatus, 96-well plates, Inverted microscope.
Procedure:
- Larval Preparation: Isolate eggs from female A. suum worms and embryonate them. Hatch the embryonated eggs in HBSS with antibiotics at 37°C and 5% CO₂ to obtain L3 larvae. Separate larvae using a Baermann apparatus and collect active larvae.
- Assay Setup: Transfer approximately 50 live, motile larvae per well into a 96-well plate containing RPMI-1640 medium.
- Compound Treatment: Add magnolol (in DMSO, final DMSO concentration ≤1%) to the wells across a range of concentrations (e.g., 1-100 µM). Include a negative control (vehicle only) and a positive control (Ivermectin).
- Incubation and Assessment: Incubate the plate at 37°C and 5% CO₂ for 24 hours. After incubation, count the number of live larvae (defined as those exhibiting movement within 4 seconds of observation) in each well under a microscope.
- Data Calculation: Calculate larval mortality for each concentration using the formula:
Mortality (%) = [1 - (Live larvae 24h after treatment / Live larvae before treatment)] × 100% - Dose-Response Analysis: Plot mortality (%) against the log of compound concentration and calculate the EC₅₀ value using non-linear regression.
Notes: This assay confirmed magnolol's anthelmintic effect with an EC₅₀ of 11.08 µM, primarily through inhibition of the mitochondrial electron transport chain [7].
Magnolol represents a multifaceted therapeutic agent with demonstrated efficacy in oncology, immunology, and parasitology. Its ability to target critical pathways like EGFR/NF-κB, induce mitochondrial dysfunction, and modulate the immune response underscores its significant potential [1] [6] [7]. However, inherent limitations of the native compound, such as poor solubility and bioavailability, necessitate advanced strategies. The development of semi-synthetic derivatives, particularly mitochondrion-targeted compounds like MTP, and the application of synthetic biology for its biosynthesis are promising avenues to enhance its therapeutic index and enable scalable production [5] [3] [4]. Future research should focus on advancing these derivatives through preclinical and clinical trials, further optimizing synthetic biology platforms for high-yield magnolol production, and exploring its synergistic potential with existing therapies across a broader range of diseases.
Magnolol, a bioactive lignan found primarily in the bark of Magnolia officinalis, has garnered significant scientific interest due to its diverse pharmacological properties, including antimicrobial, anti-inflammatory, and anticancer activities [8] [9]. The growing demand for magnolol in pharmaceutical and cosmetic applications is severely constrained by the limitations of its traditional sourcing methods. This application note details these critical bottlenecks and provides the research community with established protocols for quantifying the challenges and exploring synthetic biology solutions.
Quantitative Analysis of Traditional Sourcing Limitations
The constraints of conventional magnolol production can be summarized through the following key data points, which highlight the inefficiency and unsustainability of current practices.
Table 1: Key Limitations in Traditional Magnolol Production
| Limitation Factor | Quantitative Metric | Impact and Context |
|---|---|---|
| Cultivation Time [5] | 10 to 15 years | The lengthy growth period required for Magnolia officinalis trees before bark harvest creates a significant supply bottleneck and impedes rapid response to market demands. |
| Final Product Concentration [5] | ~1% (in dry bark) | The low concentration of magnolol in the plant material makes extraction processes inherently inefficient and resource-intensive. |
| Chemical Synthesis Challenge [5] | Low yield, multiple by-products | Traditional chemical synthesis routes are hampered by non-specific reactions, leading to poor yields and generation of undesirable by-products, complicating purification. |
Experimental Protocols for Validation and Analysis
To empirically characterize these limitations and advance alternative production platforms, the following laboratory protocols are essential.
Protocol: Quantification of Magnolol in Plant Material via HPLC
Objective: To accurately determine the concentration of magnolol in dried bark of Magnolia officinalis. Background: This protocol validates the low yield of magnolol from its natural source, providing a baseline for evaluating alternative production methods [5].
Materials & Reagents:
- Dried, powdered bark of Magnolia officinalis
- HPLC-grade methanol, acetonitrile, and purified water
- Analytical standard of magnolol (purity >98%)
- Ultrasonic water bath
- Vacuum filtration setup
- High-Performance Liquid Chromatography (HPLC) system with a UV detector
Procedure:
- Extraction: Precisely weigh 1.0 g of powdered bark. Add 10 mL of methanol and sonicate in a water bath for 60 minutes at 40°C.
- Filtration: Centrifuge the extract and filter the supernatant through a 0.45 μm membrane filter.
- HPLC Analysis:
- Column: C18 column (e.g., 150 mm x 4.6 mm, 3 μm)
- Mobile Phase: Utilize a gradient of solvent A (water with 0.1% formic acid) and solvent B (acetonitrile with 0.1% formic acid).
- Gradient Program: 0 min: 50% B; 10 min: 80% B; 15 min: 100% B; hold for 5 min.
- Flow Rate: 0.8 mL/min
- Detection: UV absorbance at 290 nm
- Injection Volume: 10 μL
- Quantification: Generate a calibration curve using the magnolol standard at concentrations of 5, 10, 25, 50, and 100 μg/mL. Calculate the magnolol concentration in the bark sample from the curve.
Protocol: In Vitro Biosynthesis of Magnolol Using Recombinant Laccase
Objective: To demonstrate the one-step enzymatic conversion of chavicol to magnolol, establishing a foundational reaction for synthetic biology approaches [5]. Background: This protocol outlines a key reaction for a potential synthetic biology route to magnolol, bypassing the need for plant cultivation.
Materials & Reagents:
- Recombinant MoLAC14 laccase (or other functional ortholog)
- Chavicol substrate
- Sodium acetate buffer (100 mM, pH 5.0)
- Orbital shaker incubator
- HPLC system (as in Protocol 3.1)
Procedure:
- Reaction Setup: In a 2 mL microcentrifuge tube, combine 970 μL of sodium acetate buffer, 20 μL of a 50 mM chavicol stock solution (in DMSO, final concentration 1 mM), and 10 μL of purified MoLAC14 laccase.
- Incubation: Incubate the reaction mixture in an orbital shaker at 30°C and 200 rpm for 4 hours.
- Reaction Termination: Stop the reaction by adding 1 mL of ice-cold methanol.
- Analysis: Centrifuge the mixture and analyze the supernatant by HPLC using the method described in Protocol 3.1 to detect and quantify the formation of magnolol.
Pathway and Workflow Visualization
The logical and experimental workflow for addressing the sourcing limitations is summarized in the following diagrams.
Diagram 1: R&D strategy for magnolol production.
The proposed biosynthetic pathway for magnolol in Magnolia officinalis, which serves as the blueprint for synthetic biology efforts, is as follows:
Diagram 2: Proposed magnolol biosynthetic pathway.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Magnolol Biosynthesis Research
| Research Reagent | Function/Application | Key Details |
|---|---|---|
| Laccase MoLAC14 [5] | Key enzyme catalyzing the dimerization of chavicol to form magnolol. | Critical for synthetic biology routes. Engineered variants (e.g., E345P, L532A) show improved thermal stability and activity. |
| Chavicol [5] | Direct precursor substrate for magnolol synthesis in the proposed enzymatic pathway. | Serves as the starting material for the one-step in vitro biosynthesis of magnolol using laccase. |
| Magnolol Standard | Essential analytical standard for quantification and method calibration. | Used in HPLC analysis (e.g., Protocol 3.1) to identify and quantify magnolol in extracts or biosynthesis reaction mixtures. |
| HPLC System with C18 Column | Core analytical platform for separation, identification, and quantification of magnolol and related compounds. | Enables quality control and yield determination for both traditional extraction and novel synthesis methods. |
| 14,15-Leukotriene A4 Methyl Ester | 14,15-Leukotriene A4 Methyl Ester, MF:C21H32O3, MW:332.5 g/mol | Chemical Reagent |
| G-quadruplex ligand 1 | G-quadruplex ligand 1, MF:C40H50N8O3, MW:690.9 g/mol | Chemical Reagent |
Within the framework of synthetic biology approaches for the production of the valuable natural product magnolol, elucidating its native biosynthetic pathway in Magnolia officinalis is a fundamental prerequisite. Magnolol, a hydroxylated biphenyl compound with potent antibacterial, anti-inflammatory, and anti-cancer properties, is traditionally extracted from the bark of Magnolia officinalis [8] [3]. However, direct plant extraction faces significant challenges due to the long cultivation time required (10–15 years) and the low concentration of magnolol in the plant (approximately 1%) [10]. Synthetic biology offers a promising alternative for the sustainable manufacturing of magnolol, but this requires a complete understanding of its biosynthetic genes and enzymes [10] [11]. For decades, the plant-based biosynthesis of magnolol remained obscure [10]. While it was speculated to stem from the common lignan biosynthesis pathway, recent research has identified a more direct route involving the coupling of two chavicol molecules [10] [12]. This application note details the identification and validation of the one-step conversion of chavicol to magnolol, catalyzed by a specific laccase enzyme, MoLAC14, and outlines experimental protocols for its study and optimization [10].
The Chavicol-to-Magnolol Biosynthetic Pathway
Pathway Hypothesis and Key Enzymes
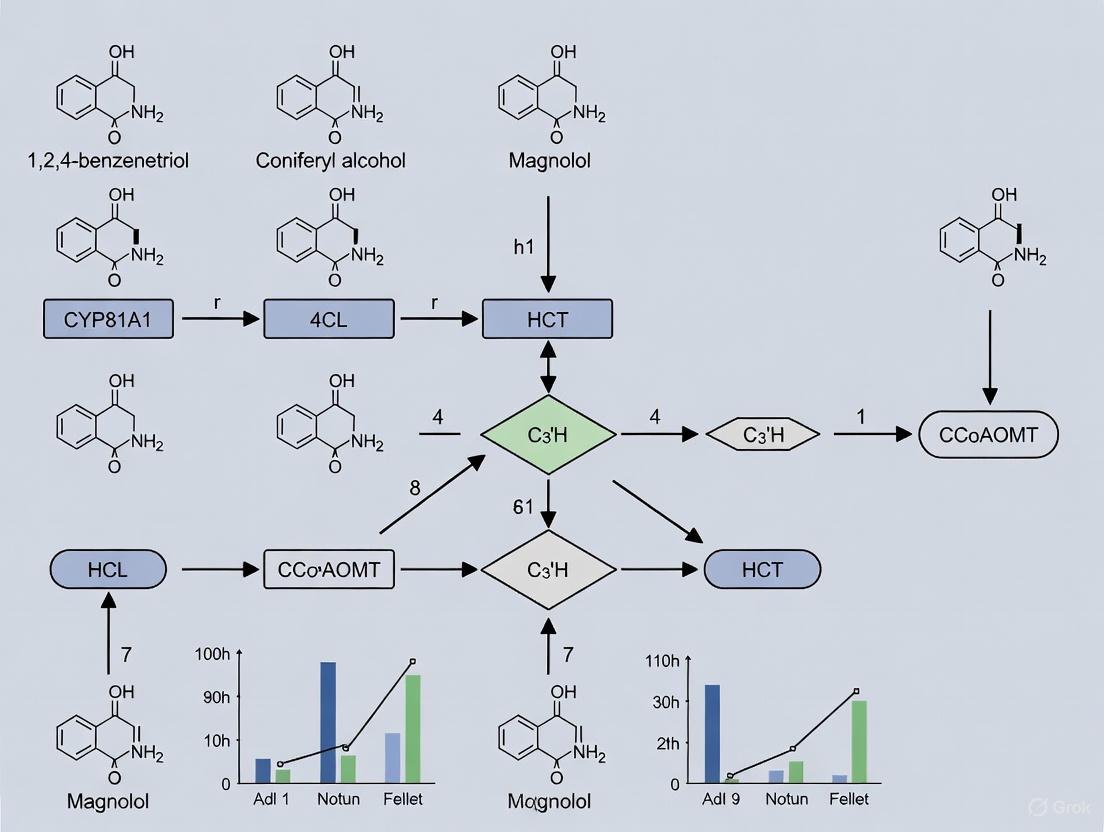
The proposed biosynthetic pathway for magnolol begins with the amino acid tyrosine. Through a series of enzymatic steps involving tyrosine ammonia-lyase (TAL), 4-coumarate CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), and alcohol dehydrogenase (ADH), tyrosine is converted to p-coumaryl alcohol [10]. Subsequent action by coniferyl alcohol acetyltransferase (CAAT) and allylphenol synthases (APS) yields the immediate precursor, chavicol [10]. The critical, one-step conversion of magnolol from chavicol is hypothesized to be catalyzed by an oxidative enzyme, laccase [10]. Laccases oxidize phenolic substrates, generating dimeric products through the coupling of radical intermediates. In this case, the oxidative coupling of two chavicol molecules forms the magnolol biphenyl structure [10] [12].
Identification of the Key Laccase Gene, MoLAC14
To validate this hypothesized pathway, a comprehensive transcriptome analysis of various tissues from Magnolia officinalis was conducted [10]. This effort identified 30 potential laccase genes in the M. officinalis genome. Gene expression analysis revealed high expression of specific laccases in roots, leaves, and bark tissues. Two gene clusters, MoLAC4 and MoLAC17, were identified as potential contributors to magnolol biosynthesis [10]. From these clusters and other highly expressed genes, six candidate laccase genes (MoSKU5F, MoLAC7B, MoLAC14, MoLAC4A, MoLAC4B, and MoLAC17F) were selected for functional characterization [10]. In vitro enzymatic assays confirmed that MoLAC14 is the pivotal enzyme responsible for the conversion of chavicol into magnolol [10].
Experimental Protocols
Protocol 1: In Vitro Assay for Laccase Activity and Magnolol Production
Objective
To express and purify candidate laccase enzymes and test their ability to catalyze the conversion of chavicol to magnolol in a controlled in vitro system [10].
Materials and Reagents
Table 1: Key Research Reagent Solutions for Laccase Assays
| Reagent / Material | Function / Description | Source / Example |
|---|---|---|
| pET-28a Vector | Expression vector for cloning and expressing laccase genes in E. coli. | Standard laboratory supplier |
| BL21(DE3) E. coli | Bacterial strain for protein expression. | Standard laboratory supplier |
| Chavicol | The enzymatic substrate, precursor to magnolol. | Chemical supplier (e.g., Sigma-Aldrich) |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Chemical inducer for gene expression. | Standard laboratory supplier |
| Copper (II) ions (Cu²âº) | Essential cofactor for laccase enzyme activity. | Standard laboratory supplier |
Procedure
- Gene Cloning and Expression:
- The coding sequences (CDS) of the candidate laccase genes (e.g., MoLAC14) are cloned into the pET-28a expression vector between the Nde I and Xho I restriction sites [10].
- The constructed plasmids are transformed into the BL21(DE3) E. coli expression strain [10].
- The transformed strain is cultured in a suitable medium (e.g., LB broth) at 37°C until the optical density at 600 nm (OD₆₀₀) reaches approximately 0.6 [10].
- Gene expression is induced by adding IPTG. Concurrently, Copper (II) ions (Cu²âº) are added to the culture to ensure proper folding and activity of the laccase enzyme [10].
- The culture is incubated further to allow protein production.
- Enzyme Preparation:
- Cells are harvested by centrifugation and lysed to release the soluble proteins.
- The recombinant laccase enzyme can be used in crude lysate or purified via affinity chromatography (e.g., His-tag purification) for more precise assays [10].
- In Vitro Enzymatic Reaction:
- The reaction mixture is set up containing the purified laccase enzyme and the substrate, chavicol, in an appropriate buffer [10].
- The reaction is incubated at a defined temperature and pH optimal for laccase activity.
- The reaction is stopped at various time points by heat inactivation or acidification.
- Product Analysis:
- The reaction products are analyzed using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS) [10].
- The formation of magnolol is confirmed by comparing its retention time and mass spectrum with an authentic standard.
Protocol 2: Protein Engineering to Enhance MoLAC14 Performance
Objective
To improve the thermal stability and catalytic activity of MoLAC14 through site-directed mutagenesis for more efficient magnolol production [10].
Procedure
- Mutagenesis Design:
- Based on structural analysis or sequence alignment with stable laccases, target specific amino acid residues for mutation.
- To enhance thermal stability, introduce proline residues (e.g., E345P, G377P) to rigidify flexible loops, or engineer additional disulfide bonds (e.g., E346C) [10].
- To probe and improve activity, perform alanine scanning mutagenesis to identify essential residues. Subsequently, target non-essential residues for beneficial mutations (e.g., L532A) [10].
- Site-Directed Mutagenesis:
- Introduce the desired mutations into the MoLAC14 gene in the pET-28a plasmid using a commercial mutagenesis kit or overlap extension PCR.
- Expression and Purification:
- Express and purify the mutant enzymes following the same protocol as for the wild-type enzyme (Protocol 1, steps 1-2).
- Characterization of Mutants:
- Thermal Stability Assay: Incubate wild-type and mutant enzymes at elevated temperatures for a set duration. Measure the residual activity using a standard laccase substrate (e.g., ABTS) or chavicol. Calculate the half-life or melting temperature (Tₘ) [10].
- Enzymatic Activity Assay: Measure the initial reaction rates of wild-type and mutant enzymes with chavicol as a substrate under identical conditions. Quantify magnolol production via HPLC to determine the specific activity [10].
Key Experimental Data and Findings
Validation of MoLAC14 Activity
In vitro experiments confirmed that MoLAC14 successfully catalyzes the formation of magnolol from chavicol. The product was unequivocally identified by HPLC and MS analysis, establishing MoLAC14 as the key biosynthetic enzyme in the native pathway [10].
Performance of MoLAC14 Mutants
Protein engineering led to significant improvements in the enzyme's properties. The table below summarizes the effects of key mutations on thermal stability and magnolol production yield [10].
Table 2: Effects of MoLAC14 Mutations on Enzyme Performance
| Mutation | Type of Modification | Effect on Enzyme | Impact on Magnolol Production |
|---|---|---|---|
| E345P, G377P | Introduction of proline | Enhanced thermal stability | Not Specified |
| H347F, E346F | Aromatic substitution | Enhanced thermal stability | Not Specified |
| E346C | Potential disulfide bond | Enhanced thermal stability | Not Specified |
| L532A | Alanine scan mutation | Likely enhanced catalytic activity | Boosted production to 148.83 mg/L |
The identification of MoLAC14 as the laccase catalyzing the direct conversion of chavicol to magnolol represents a critical breakthrough in elucidating the native biosynthetic route of this valuable compound [10]. Furthermore, the successful engineering of MoLAC14 to enhance its stability and the subsequent dramatic increase in magnolol titer to 148.83 mg/L demonstrates the power of enzyme optimization for metabolic engineering [10]. These findings provide the essential genetic and enzymatic foundation for the synthetic biology-driven production of magnolol. The genes encoding the enzymes from tyrosine to chavicol, combined with the optimized MoLAC14 gene, can now be assembled into a microbial chassis (e.g., E. coli or yeast) to create a cell factory for the sustainable and efficient production of magnolol, overcoming the limitations associated with traditional plant extraction [10].
Within the framework of synthetic biology for the production of magnolol, a plant-derived compound with potent antibacterial properties, the identification and engineering of key biosynthetic enzymes is a fundamental research objective [10] [11]. Traditional extraction of magnolol from the bark of Magnolia officinalis is inefficient, requiring 10–15 years of tree cultivation and yielding low concentrations of the target compound (approximately 1%) [10]. Synthetic biology offers a promising alternative, but its application has been hampered by a lack of understanding of the magnolol biosynthesis pathway in the plant [10]. Recent research has identified a one-step conversion of the precursor chavicol into magnolol, catalyzed by laccase enzymes, with the laccase MoLAC14 from M. officinalis emerging as a pivotal biocatalyst [10] [11]. This application note details the functional identification and validation protocols for MoLAC14, providing a methodological foundation for leveraging this enzyme in synthetic microbial factories for magnolol production.
Laccase Structure and Catalytic Mechanism in Magnolol Synthesis
Laccases (EC 1.10.3.2) are multi-copper oxidases that catalyze the one-electron oxidation of a broad range of substrates, including phenolics, with the concomitant four-electron reduction of molecular oxygen to water [13] [14]. This makes them ideal green catalysts for synthetic biology, as they operate without the need for cofactors and produce water as the only by-product [14] [15].
The catalytic center of laccase contains four copper atoms, classified into three types: a Type 1 (T1) copper, which is responsible for the enzyme's characteristic blue color and is the primary site for substrate oxidation; a Type 2 (T2) copper; and a binuclear Type 3 (T3) copper cluster [13] [15]. The T2 and T3 coppers form a trinuclear cluster (TNC) where the reduction of oxygen occurs. The proposed mechanism for magnolol synthesis involves the oxidation of two chavicol molecules by the T1 copper. The electrons removed from the substrates are transferred internally via a conserved His-Cys-His tripeptide bridge to the trinuclear cluster, where oxygen is reduced to water. The oxidized chavicol molecules form radicals that subsequently couple to form magnolol [10] [14]. The following diagram illustrates this catalytic process and its integration into the experimental workflow for identifying functional laccases.
Identification and Validation of MoLAC14
Transcriptome Analysis and Candidate Gene Selection
The identification of MoLAC14 began with a comprehensive transcriptomic analysis of various tissues from M. officinalis [10]. This involved:
- RNA Sequencing: Transcriptome sequencing of 20 samples from different tissues (e.g., bark, leaves, roots) of 16-year-old M. officinalis plants.
- Genome Annotation Enhancement: Assembled RNA-seq reads were mapped to the existing M. officinalis genome, significantly enriching its annotation and leading to the identification of 30 distinct laccase genes [10].
- Candidate Selection: Two potential laccase gene clusters (MoLAC4 and MoLAC17) associated with magnolol production were identified. Six highly expressed laccase genes from these clusters and other magnolol-producing tissues (including MoSKU5F, MoLAC7B, MoLAC14, MoLAC4A, MoLAC4B, and MoLAC17F) were selected for functional characterization [10].
Experimental Protocol: Cloning, Expression, and In Vitro Assay
The following protocol details the key steps for the heterologous expression and functional testing of candidate laccases like MoLAC14.
Protocol 1: Functional Testing of Recombinant Laccase Activity
- Objective: To heterologously express candidate laccase genes and test their ability to catalyze the conversion of chavicol to magnolol in vitro.
- Principle: The coding sequence (CDS) of the laccase gene is cloned into an expression vector, transformed into E. coli, and the expressed enzyme is used in a reaction with chavicol. The formation of magnolol is confirmed using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS) [10].
Materials and Methods:
- Gene Synthesis and Cloning:
- The CDS of the selected laccase genes are synthesized and integrated into a pET-28a vector between the Nde I and Xho I restriction sites using Gibson assembly [10].
- Heterologous Expression:
- The constructed plasmids are chemically transformed into E. coli BL21(DE3) expression strain.
- The transformed strain is cultured in LB medium at 37°C.
- Gene expression is induced by adding Isopropyl β-D-1-thiogalactopyranoside (IPTG).
- Concurrently, Copper (II) ions (Cu²âº) are added to the culture to facilitate the proper assembly of the laccase's multi-copper center [10].
- In Vitro Activity Assay:
- The expressed laccase is purified or used in a crude cell lysate.
- The enzyme is incubated with the substrate chavicol in a suitable reaction buffer.
- Product Detection and Validation:
- The reaction mixture is analyzed using HPLC to separate and quantify the products.
- The identity of magnolol is confirmed by Mass Spectrometry (MS) by comparing its mass and fragmentation pattern to an authentic standard [10].
Results and Validation: In vitro experiments confirmed that MoLAC14 could efficiently catalyze the one-step conversion of chavicol to magnolol. HPLC and MS analyses provided definitive evidence of magnolol production, identifying MoLAC14 as a pivotal enzyme in the magnolol biosynthetic pathway [10] [11].
Engineering MoLAC14 for Enhanced Performance
Protein engineering was employed to enhance the thermal stability and catalytic efficiency of MoLAC14, which is critical for its application in industrial bioprocesses.
Protocol 2: Engineering MoLAC14 for Improved Stability and Yield
- Objective: To improve the thermal stability and catalytic activity of MoLAC14 through site-directed mutagenesis.
- Principle: Targeted mutations are introduced into the MoLAC14 gene to stabilize the protein structure or alter active site residues, followed by functional screening of the variants [10] [16].
Materials and Methods:
- Site-Directed Mutagenesis:
- Mutations are introduced into the MoLAC14 gene sequence using techniques such as PCR-based site-directed mutagenesis.
- Key mutations that were experimentally validated include:
- Stability Mutations: E345P, G377P, H347F, E346C, E346F. These proline and residue substitutions were designed to enhance the rigidity and thermal stability of the enzyme [10].
- Activity Mutations: Alanine scanning was performed to identify essential residues. The mutation L532A was found to significantly boost magnolol production [10].
- Expression and Purification:
- The mutant genes are cloned and expressed in E. coli BL21(DE3) following Protocol 1.
- The mutant enzymes are purified for biochemical characterization.
- Biochemical Characterization:
- Thermal Stability Assay: The wild-type and mutant enzymes are incubated at elevated temperatures for a set period, and the residual activity is measured and compared.
- Activity Assay: Enzyme activity is measured by monitoring the rate of magnolol production from chavicol under standardized conditions.
Results and Performance Data: The engineered MoLAC14 variants showed marked improvements over the wild-type enzyme. The table below summarizes the quantitative enhancements achieved through protein engineering.
Table 1: Enhanced Performance of Engineered MoLAC14 Variants
| Mutation | Impact on Enzyme Properties | Reported Outcome |
|---|---|---|
| E345P, G377P, H347F, E346C, E346F | Notably enhanced thermal stability [10]. | Improved enzyme stability under process conditions. |
| L532A | Boosted magnolol production yield [10]. | Increased magnolol titer to an unprecedented level of 148.83 mg/L [10]. |
The Scientist's Toolkit: Research Reagent Solutions
The table below lists key reagents and materials essential for the functional identification and characterization of laccases like MoLAC14, based on the protocols described.
Table 2: Essential Research Reagents for Laccase Identification and Assay
| Reagent / Material | Function / Application | Specific Example / Note |
|---|---|---|
| pET-28a Vector | Expression vector for heterologous protein production in E. coli [10]. | Used for cloning MoLAC14 and its variants. |
| E. coli BL21(DE3) | A robust host strain for recombinant protein expression [10] [16]. | Chemically transformed with the expression plasmid. |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Inducer for triggering the expression of the target gene in the pET system [10] [16]. | Added to the bacterial culture to initiate laccase production. |
| Copper (II) Sulfate (CuSO₄) | Source of Cu²⺠ions essential for the assembly and function of the laccase multi-copper center [10] [16]. | Added to the culture medium during induction. |
| Chavicol | The phenolic substrate for the laccase-catalyzed reaction in magnolol synthesis [10]. | Converted to magnolol by MoLAC14. |
| ABTS (2,2'-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) | A common synthetic substrate used for high-throughput screening of laccase activity [16]. | Oxidation produces a colored product, easy to monitor. |
| HPLC-MS System | Analytical platform for separating, detecting, and confirming the identity of magnolol from reaction mixtures [10]. | Critical for validating the enzymatic function. |
| Monomethyl auristatin E intermediate-17 | Monomethyl auristatin E intermediate-17, MF:C27H35NO7S, MW:517.6 g/mol | Chemical Reagent |
| Cyromazine-3-mercaptopropanoic acid | Cyromazine-3-mercaptopropanoic Acid|Research Grade | Cyromazine-3-mercaptopropanoic acid is a research chemical for laboratory investigation. This product is For Research Use Only. Not for human or veterinary use. |
The functional identification and subsequent engineering of the laccase MoLAC14 represent a significant advancement in the synthetic biology of magnolol production. The detailed protocols for transcriptome-driven gene discovery, heterologous expression, in vitro functional assays, and protein engineering provide a robust roadmap for researchers. The successful enhancement of MoLAC14's stability and yield through rational design underscores the potential of enzyme engineering in creating efficient biocatalysts. Integrating this optimized enzyme into engineered microbial hosts promises a sustainable and scalable manufacturing route for magnolol, overcoming the limitations of traditional plant extraction.
The identification of genes within biosynthetic pathways is a critical step in synthetic biology, particularly for the production of valuable plant-derived compounds like magnolol. Magnolol, a bioactive lignan from Magnolia officinalis, exhibits potent antibacterial, anti-inflammatory, and neuroprotective properties but faces production challenges due to the plant's long cultivation time and low compound concentration [10] [8]. Traditional extraction and chemical synthesis methods are often inefficient, environmentally unsustainable, and produce low yields [8]. Synthetic biology offers a promising alternative, but its application requires a deep understanding of the underlying biosynthetic genes and pathways [10]. This application note details integrated genomic and transcriptomic protocols for discovering key genes, using the identification of the magnolol biosynthesis enzyme MoLAC14 as a case study. These methodologies enable researchers to move from raw multi-omics data to functionally validated genes, paving the way for the microbial production of magnolol and other high-value natural products.
Experimental Protocols
Protocol 1: Transcriptome Sequencing and Assembly for Pathway Hypothesis Generation
Principle: Comprehensive transcriptome sequencing across diverse plant tissues provides the foundational data to identify genes involved in specialized metabolism. This protocol focuses on generating a high-quality transcriptome assembly to probe the proposed magnolol biosynthetic pathway starting from chavicol [10].
Procedure:
- Tissue Collection: Collect multiple biological replicates (e.g., n=3-4) from various tissues (e.g., bark, leaves, roots) of Magnolia officinalis. Tissue selection should be informed by the known accumulation pattern of the target compound.
- RNA Extraction and QC: Extract total RNA using a standard kit (e.g., Qiagen RNeasy Plant Mini Kit). Assess RNA integrity and purity using an Agilent Bioanalyzer, ensuring all samples have an RNA Integrity Number (RIN) > 8.0.
- Library Preparation and Sequencing: Prepare strand-specific RNA-seq libraries (e.g., using Illumina TruSeq Stranded mRNA kit). Sequence the libraries on an Illumina platform (e.g., NovaSeq 6000) to a minimum depth of 6 Gb of high-quality (Q30 > 97%) data per sample [10].
- Read Processing and Assembly: Trim raw reads to remove adapters and low-quality bases using Trimmomatic. Map the high-quality reads to a reference genome of M. officinalis (if available) using HISAT2 or STAR. For de novo assembly, use Trinity software to reconstruct transcripts without a reference genome.
- Transcriptome Annotation: Perform functional annotation of the assembled transcripts using BLAST against databases like UniProt and Swiss-Prot, and assign Gene Ontology (GO) terms using tools like InterProScan.
Troubleshooting: Low mapping rates may indicate poor RNA quality or genomic divergence; consider de novo assembly. Incomplete BUSCO scores suggest an incomplete transcriptome; leverage multiple assembly algorithms and combine results.
Protocol 2: Identification and Phylogenetic Analysis of Candidate Gene Families
Principle: Based on a biosynthetic hypothesis (e.g., laccase-mediated dimerization of chavicol to form magnolol), this protocol details the process of identifying all members of a target gene family and prioritizing candidates for functional testing [10].
Procedure:
- Family-Specific HMMER Search: Using hidden Markov models (HMMs) for the protein domain of interest (e.g., laccase, PFAM domain PF00394), search the annotated M. officinalis proteome using HMMER3 to identify all putative family members.
- Differential Expression Analysis: Calculate transcripts per million (TPM) values for each gene across all sequenced tissues. Perform differential expression analysis using DESeq2 or edgeR to identify genes highly expressed in tissues known to produce magnolol.
- Identification of Gene Clusters: Examine the genomic locations of the identified candidate genes. Genes arranged in tandem within a 100-200 Kb genomic region are considered part of a biosynthetic gene cluster. Identify such clusters as they are often linked to secondary metabolite pathways [10].
- Phylogenetic Analysis: Align the protein sequences of the candidate genes with homologs from model plants (e.g., Arabidopsis thaliana) using MUSCLE. Construct a phylogenetic tree with IQ-TREE using maximum likelihood. Candidates that form species-specific clades or expand via tandem duplication are high-priority targets.
Troubleshooting: A large number of candidates can be prioritized by integrating expression levels (e.g., TPM > 50 in productive tissues) and phylogenetic clustering.
Protocol 3: Heterologous Expression and In Vitro Enzyme Activity Assay
Principle: This protocol validates the function of a candidate gene by heterologously expressing it in E. coli and testing its ability to convert a proposed substrate (chavicol) into the desired product (magnolol) in a controlled in vitro environment [10].
Procedure:
- Gene Synthesis and Cloning: Synthesize the codon-optimized coding sequence (CDS) of the candidate gene (e.g., MoLAC14). Clone the CDS into a protein expression vector (e.g., pET-28a) between NdeI and XhoI restriction sites using Gibson Assembly, resulting in an N-terminal His-tag fusion [10].
- Transformation and Expression: Chemically transform the constructed plasmid into E. coli BL21(DE3) cells. Grow a culture in LB medium at 37°C until OD600 reaches 0.6-0.8. Induce protein expression with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and add 0.5 mM CuSO4 to the medium to facilitate laccase metallation. Incubate for 16-20 hours at 16°C.
- Protein Purification: Harvest cells by centrifugation and lyse via sonication. Purify the soluble His-tagged protein using Ni-NTA affinity chromatography. Elute the protein with an imidazole gradient and desalt into an appropriate reaction buffer (e.g., 50 mM phosphate buffer, pH 7.0).
- In Vitro Enzyme Assay: In a reaction mixture, combine purified enzyme (e.g., 10 µg), substrate (chavicol, 1 mM), and buffer. Incubate at 30°C for 1-2 hours.
- Product Analysis by HPLC/MS: Stop the reaction by adding an equal volume of methanol. Analyze the metabolites via High-Performance Liquid Chromatography (HPLC) coupled with Mass Spectrometry (MS). Identify magnolol by comparing its retention time and mass signature to an authentic standard.
Troubleshooting: Low protein solubility may require optimization of induction temperature and IPTG concentration. Lack of activity may indicate improper cofactor incorporation or the need for different reaction conditions (e.g., pH, oxygen availability).
Protocol 4: Enzyme Engineering for Enhanced Stability and Production
Principle: Once a key enzyme is identified, its properties can be improved through rational engineering. This protocol uses site-directed mutagenesis to enhance thermal stability and enzymatic activity, thereby increasing final product titers [10].
Procedure:
- Rational Mutagenesis Design: Based on structural models or homology to stable laccases, design mutations (e.g., E345P, G377P) to introduce stabilizing prolines or mutate residues in the substrate-binding pocket (e.g., L532A) [10].
- Site-Directed Mutagenesis: Use a commercial kit (e.g., Q5 Site-Directed Mutagenesis Kit, NEB) to introduce the desired mutations into the expression plasmid, following the manufacturer's instructions.
- Expression and Purification: Express and purify the mutant enzymes following steps 2-4 of Protocol 3.
- Thermal Stability Assay: Use a thermal shift assay (e.g., with Sypro Orange dye) to measure the melting temperature (Tm) of wild-type and mutant enzymes. An increase in Tm indicates improved stability.
- Fermentation and Titer Measurement: Express the best-performing mutant enzyme in a production host (e.g., engineered S. cerevisiae or E. coli) in a bioreactor. Quantify magnolol production in the culture medium over time using HPLC. The L532A mutation in MoLAC14 has been shown to boost magnolol production to 148.83 mg/L [10].
Troubleshooting: Some mutations may abolish activity; always screen multiple clones and characterize a range of mutations (e.g., via alanine scanning).
Data Presentation and Analysis
Quantitative Data from MoLAC14 Engineering
The following table summarizes the quantitative outcomes of engineering the key magnolol biosynthesis enzyme, MoLAC14, demonstrating the impact of specific mutations on thermal stability and product yield [10].
Table 1: Impact of MoLAC14 Mutations on Enzyme Properties and Magnolol Production
| Mutation | Property Targeted | Key Outcome | Reported Magnolol Titer |
|---|---|---|---|
| E345P, G377P | Thermal Stability | Enhanced stability | Not Specified [10] |
| H347F, E346C, E346F | Thermal Stability | Enhanced stability | Not Specified [10] |
| L532A | Enzymatic Activity | Boosted catalytic efficiency | 148.83 mg/L [10] |
| Wild-type MoLAC14 | (Baseline) | Baseline activity | Not Specified [10] |
Research Reagent Solutions
The following table catalogues essential reagents and tools for executing the gene discovery and validation workflows described in this application note.
Table 2: Key Research Reagent Solutions for Omics-Driven Gene Discovery
| Reagent / Tool | Function / Application | Example / Specification |
|---|---|---|
| RNA-seq Library Prep Kit | Preparation of strand-specific RNA sequencing libraries. | Illumina TruSeq Stranded mRNA Sample Preparation Kit |
| Sequence Assembly Software | De novo transcriptome assembly from RNA-seq reads. | Trinity (v2.15.1) |
| Protein Expression System | Heterologous expression and purification of candidate enzymes. | pET-28a vector in E. coli BL21(DE3); Ni-NTA Resin |
| Chromatography System | Separation, identification, and quantification of reaction products. | HPLC System with C18 column coupled to a Q-TOF Mass Spectrometer |
| Multi-omics Integration Tool | Computational integration of genomic, transcriptomic, and pharmacological data. | MiDNE (Multi-omics genes and Drugs Network Embedding) R package [17] |
Pathway and Workflow Visualizations
Proposed Magnolol Biosynthesis Pathway
The following diagram illustrates the hypothesized biosynthetic pathway from chavicol to magnolol in Magnolia officinalis, culminating in the laccase-catalyzed coupling step validated in this study [10].
Proposed Biosynthesis Pathway
Gene Discovery and Validation Workflow
This diagram outlines the complete experimental workflow from tissue sampling and omics data generation to the final functional validation and engineering of the candidate gene.
Gene Discovery Workflow
Building Cell Factories: Engineering Microbial Hosts and Biosynthesis Pathways
Synthetic biology provides a powerful framework for engineering cellular factories to produce high-value natural compounds like magnolol, a potent antibacterial plant-derived molecule [5]. Central to this endeavor is the selection of an appropriate microbial chassis—a host organism engineered to carry out a desired biological function. The ideal chassis determines the efficiency, yield, and scalability of the entire bioprocess. Escherichia coli and various yeast species, particularly Saccharomyces cerevisiae, stand as the foundational workhorses in this field due to their well-characterized genetics, rapid growth, and the extensive availability of synthetic biology tools [18]. This article examines the prospects of these and other chassis organisms, providing a structured comparison and detailed experimental protocols, all framed within the context of optimizing magnolol production.
Comparative Analysis of Microbial Chassis Organisms
Selecting a host organism involves balancing factors such as growth rate, ability to perform complex modifications, yield, and cost. The table below summarizes the key characteristics of the most commonly used microbial systems.
Table 1: Key Features of Promising Microbial Chassis Organisms
| Aspect | Escherichia coli | Saccharomyces cerevisiae (Yeast) | Pichia pastoris (Yeast) | Bacillus subtilis |
|---|---|---|---|---|
| Key Advantages | Rapid growth, easy genetic manipulation, low cost, extensive molecular tool availability [19] [20] | Performs eukaryotic post-translational modifications (PTMs), well-established engineering toolkit, robust [18] [20] | High cell density fermentation, performs glycosylation, strong inducible promoters [20] | Naturally secretes proteins, GRAS status, simplified purification [20] |
| Key Limitations | Limited PTMs, formation of inclusion bodies, lower tolerance to some flavonoids [19] [21] | Longer doubling time than bacteria, more complex genetics, higher cost than bacterial systems [19] [20] | Requires precise optimization, higher cost compared to bacterial systems, methanol use in some systems [20] | Limited PTMs, requires strain-specific optimization for some proteins [20] |
| Post-Translational Modifications | Minimal to none [20] | Yes (e.g., glycosylation) [20] | Yes (e.g., eukaryotic-like glycosylation) [20] | Minimal to none [20] |
| Typical Protein Yield | 1-10 g/L (intracellular) [19] | Up to 20 g/L (reported for various proteins) [19] | Information not specified in search results | Information not specified in search results |
| Growth Rate | Very fast (doubling time ~20 min) [20] | Moderate (doubling time ~2 hours) [20] | Moderate | Moderate (doubling time ~30-60 min) [20] |
| Ideal Applications | Enzymes, small therapeutic proteins, simple natural products [20], flavonoid glycosylation [21] | Production of complex therapeutic proteins, secondary metabolites requiring eukaryotic PTMs [18] [19] | Therapeutic proteins, enzymes requiring glycosylation [20] | Industrial enzymes, bulk production of soluble proteins [20] |
For magnolol production, which involves the dimerization of chavicol, the choice of chassis is critical. Research has identified the laccase enzyme MoLAC14 as the key catalyst for this one-step conversion [5]. A chassis like E. coli may be sufficient if the laccase can be functionally expressed and the product does not require further eukaryotic-specific modifications. However, non-model E. coli strains can offer significant advantages. For instance, E. coli W has demonstrated enhanced tolerance to toxic flavonoids like chrysin compared to the standard K-12 strain, making it a superior platform for flavonoid glycosylation processes [21]. For more complex pathways where subcellular compartmentalization or specialized PTMs are beneficial, a yeast chassis like S. cerevisiae might be preferable.
Application Notes: Chassis Engineering for Magnolol Production
Case Study: Establishing a RobustE. coliPlatform
A recent study exemplifies the systematic engineering of a chassis for flavonoid bioprocessing. Researchers selected E. coli W for its innate resilience and ability to utilize sucrose, a low-cost carbon source. To enhance its performance, they employed a two-pronged approach:
- Adaptive Laboratory Evolution (ALE): The strain was evolved under sucrose selection pressure to optimize its native sucrose metabolism [21].
- Targeted Metabolic Engineering: Key genes (xylA, zwf, pgi) were knocked out to reroute intracellular carbon flux away from biomass and toward the synthesis of uridine diphosphate glucose (UDPG), a crucial precursor for glycosylation reactions [21]. This engineered chassis, when applied to the glycosylation of the flavonoid chrysin, achieved a remarkable titer of 1844 mg/L in a scaled-up fed-batch bioreactor [21]. This demonstrates the power of combining chassis innate properties with advanced engineering to overcome bioprocess constraints like precursor availability and product toxicity.
Protocol: In Vivo Magnolol Production in EngineeredE. coli
This protocol details the steps for producing magnolol in an engineered E. coli chassis by expressing the key laccase gene MoLAC14 from Magnolia officinalis [5].
Principle: The protocol leverages recombinant DNA technology to introduce the plant-derived MoLAC14 gene into E. coli. The engineered host then expresses the laccase enzyme, which catalyzes the oxidative coupling of two chavicol molecules to form magnolol [5].
Workflow: The following diagram illustrates the experimental workflow for the production and analysis of magnolol in an engineered E. coli system.
Materials:
- Strain: E. coli BL21(DE3) or other suitable expression host.
- Vector: pET-28a plasmid (or similar with T7/lac promoter and antibiotic resistance) [5].
- Gene: Codon-optimized MoLAC14 gene (GenBank accession number can be found in [5]).
- Media: LB broth and agar plates with appropriate antibiotic (e.g., 50 µg/mL kanamycin for pET-28a).
- Inducer: Isopropyl β-d-1-thiogalactopyranoside (IPTG).
- Substrate: Chavicol.
- Equipment: Shaking incubator, centrifuge, spectrophotometer, HPLC system, mass spectrometer.
Procedure:
- Plasmid Construction: Subclone the MoLAC14 coding sequence into the pET-28a expression vector between the NdeI and XhoI restriction sites using standard molecular biology techniques or Gibson assembly [5].
- Transformation: Introduce the constructed plasmid into chemically competent E. coli BL21(DE3) cells via heat shock transformation. Plate the cells on LB agar plates containing the appropriate antibiotic and incubate overnight at 37°C.
- Culture and Induction: Inoculate a single colony into 5 mL of LB medium with antibiotic and grow overnight at 37°C with shaking. Use this starter culture to inoculate a larger culture (e.g., 50-100 mL) at a 1:100 dilution. Grow the culture at 37°C until the OD600 reaches approximately 0.6. Add IPTG to a final concentration of 0.1-1.0 mM to induce laccase expression. Lower the temperature to 25-30°C and continue incubation for 4-16 hours.
- Biotransformation: Add chavicol (e.g., 0.5-1.0 mM) to the induced culture and incubate further for 12-24 hours to allow for enzymatic conversion to magnolol.
- Product Extraction: Harvest the cells by centrifugation (e.g., 4,000 x g for 20 minutes). Resuspend the cell pellet in methanol or ethyl acetate and vortex vigorously to extract magnolol. Separate the organic solvent phase by centrifugation.
- Analysis:
- HPLC: Analyze the extract using reverse-phase HPLC. Monitor for a peak corresponding to the magnolol standard (retention time ~15.3 minutes under conditions in [5]).
- Mass Spectrometry (MS): Confirm the identity of the product using LC-MS. Magnolol has a molecular weight of 266.1 g/mol, and the [M-H]â» ion is typically observed at m/z 265 [5].
Protocol: Engineering Enhanced Enzyme Activity for Improved Titer
A critical strategy in synthetic biology is to optimize the pathway enzymes themselves. This protocol describes the engineering of MoLAC14 for improved thermal stability and activity, which directly translates to higher magnolol production [5].
Principle: Through site-directed mutagenesis, specific amino acid residues in the laccase enzyme are altered. These mutations can enhance protein stability and catalytic efficiency, leading to a more effective whole-cell biocatalyst.
Workflow: The diagram below outlines the key steps in the enzyme engineering and validation cycle.
Materials:
- Template: Plasmid containing the wild-type MoLAC14 gene.
- Primers: Mutagenic primers designed for the target site (e.g., to introduce the L532A mutation).
- Kit: Site-directed mutagenesis kit.
- Equipment: Thermocycler, equipment for protein purification (e.g., affinity chromatography), spectrophotometer for enzyme assays, thermal block.
Procedure:
- Mutation Design: Based on structural models or alignments with stable laccases, design mutations. For MoLAC14, mutations like E345P, G377P, and L532A have been shown to improve stability and activity [5].
- Site-Directed Mutagenesis: Perform the mutagenesis reaction according to the manufacturer's protocol to create the variant MoLAC14 plasmid.
- Expression and Purification: Transform, express, and purify the wild-type and mutant enzymes as described in the previous protocol.
- Enzyme Assay: Measure laccase activity by monitoring the oxidation of a substrate like ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) at 420 nm, or directly by quantifying magnolol production from chavicol via HPLC.
- Thermal Stability Assay: Incubate the purified enzymes at a elevated temperature (e.g., 50°C) for a set time. Remove aliquots at regular intervals and measure the remaining activity. The half-life of the mutant enzyme should be compared to the wild-type.
- Whole-Cell Validation: Introduce the best-performing mutant gene into the production chassis and run the magnolol production protocol. The L532A mutation, for example, boosted magnolol production to 148.83 mg/L in a laboratory setting [5].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagent Solutions for Synthetic Biology-Driven Magnolol Research
| Research Reagent | Function/Application in Magnolol Research |
|---|---|
| pET-28a Vector | A common protein expression plasmid used for heterologous expression of the MoLAC14 laccase gene in E. coli [5]. |
| CRISPR/Cas9 System | Enables precise, scarless genome editing in chassis organisms (e.g., E. coli, yeast) for metabolic engineering, such as knocking out genes to redirect metabolic flux [18] [21] [22]. |
| Chavicol | The direct phenolic precursor molecule that is dimerized by the laccase enzyme MoLAC14 to form magnolol [5]. |
| UDP-Glucose (UDPG) | A key nucleotide sugar precursor for glycosylation reactions. Enhancing its intracellular availability is critical for producing glycosylated flavonoids, which can improve solubility and bioavailability [21]. |
| Adaptive Laboratory Evolution (ALE) | A technique used to enhance desired chassis properties, such as improved sucrose metabolism or flavonoid tolerance, by applying long-term selective pressure [21]. |
| Thalidomide-4-NH-PEG1-COO(t-Bu) | Thalidomide-4-NH-PEG1-COO(t-Bu), MF:C22H27N3O7, MW:445.5 g/mol |
| Angiotensinogen (1-13) (human) | Angiotensinogen (1-13) (human), MF:C79H116N22O17, MW:1645.9 g/mol |
The strategic selection and engineering of a microbial chassis are paramount for the successful synthetic production of magnolol. E. coli offers a fast, tractable, and cost-effective system, with non-model strains like E. coli W providing enhanced robustness for toxic compounds. Yeast chassis provide essential eukaryotic machinery for more complex engineering tasks. The decision must be guided by the specific pathway requirements, with recent advances in genome editing, enzyme engineering, and bioprocess optimization providing the necessary tools to tailor these hosts for high-value compound production. The protocols and application notes outlined here provide a concrete foundation for researchers to engineer and deploy these microbial workhorses effectively.
Within synthetic biology frameworks, the efficient production of plant-derived natural products like magnolol relies on the precise design of genetic vectors and their successful introduction into a microbial host. Magnolol, a bioactive neolignan from Magnolia officinalis, exhibits a broad range of pharmacological activities, including anti-inflammatory, anti-bacterial, and anti-parasitic effects, making it a compelling target for bioproduction [9] [7]. Traditional extraction from magnolia bark is inefficient due to long cultivation times and low compound concentration, typically around 1% [5]. Synthetic biology offers a sustainable alternative by enabling the heterologous production of such valuable compounds in engineered microorganisms. This application note details the core principles and protocols for constructing the biosynthetic machinery for magnolol production, providing a guide for researchers and scientists in drug development.
Biosynthetic Pathway and Key Enzyme Identification
The proposed biosynthetic pathway for magnolol in Magnolia officinalis begins with tyrosine. Through a series of enzymatic steps involving tyrosine ammonia-lyase (TAL), 4-coumarate CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), and alcohol dehydrogenase (ADH), p-coumaryl alcohol is produced. This intermediate is then converted to chavicol by enzymes including coniferyl alcohol acetyltransferase (CAAT) and allylphenol synthases (APS) [5].
Critically, research indicates that magnolol is synthesized from the precursor chavicol in a one-step reaction catalyzed by the enzyme laccase [5]. Leveraging transcriptome data from M. officinalis, 30 potential laccase genes were identified. Among them, MoLAC14 was functionally validated in vitro as the pivotal enzyme responsible for the oxidative coupling of two chavicol molecules to form magnolol [5].
Table 1: Key Enzymes in the Proposed Magnolol Biosynthetic Pathway
| Enzyme | Abbreviation | Function in Magnolol Biosynthesis |
|---|---|---|
| Tyrosine ammonia-lyase | TAL | Converts tyrosine to coumaric acid |
| 4-coumarate CoA ligase | 4CL | Activates coumaric acid to form coumaroyl-CoA |
| Cinnamoyl-CoA reductase | CCR | Reduces coumaroyl-CoA to cinnamaldehyde |
| Alcohol dehydrogenase | ADH | Reduces cinnamaldehyde to cinnamyl alcohol |
| Allylphenol synthase | APS | Converts p-coumaryl alcohol to chavicol |
| Laccase (MoLAC14) | LAC | Oxidatively dimerizes chavicol to form magnolol |
Vector Design and Engineering of MoLAC14
Basic Vector Construction
The coding sequence (CDS) of the target laccase gene, MoLAC14, should be cloned into an appropriate expression vector. For initial functional validation in E. coli, the following design is recommended:
- Vector Backbone: pET-28a is a suitable choice for high-level expression in E. coli [5].
- Cloning Sites: Integration of the gene between the Nde I and Xho I restriction sites.
- Assembly Method: The vector can be constructed using Gibson assembly, a method noted for its use in assembling biosynthetic gene clusters (BGCs) [5] [23].
This basic vector, pET-28a-MoLAC14, allows for inducible expression of the laccase enzyme to test its activity in converting chavicol to magnolol.
Enzyme Engineering for Enhanced Stability and Activity
Wild-type enzymes often require optimization for efficient industrial application. Engineering of MoLAC14 has demonstrated significant improvements in thermal stability and activity [5]. The following single-point mutations were identified as beneficial:
Table 2: MoLAC14 Mutations for Improved Performance
| Mutation | Impact on Enzyme Properties |
|---|---|
| E345P | Enhanced thermal stability |
| G377P | Enhanced thermal stability |
| H347F | Enhanced thermal stability |
| E346C | Enhanced thermal stability |
| E346F | Enhanced thermal stability |
| L532A | Increased magnolol production titer |
Alanine scanning, a technique for identifying essential residues, led to the discovery of the L532A mutation, which boosted magnolol production to 148.83 mg/L in a fermentation context, underscoring the critical role of protein engineering in synthetic biology [5].
Host Selection and Transformation
Chassis Selection
While E. coli is a common host for initial gene characterization and pathway validation, the choice of a production chassis is critical. For complex natural products like type II polyketides, Streptomyces species are often superior due to their native capacity for secondary metabolism and precursor supply [24]. A engineered strain of Streptomyces aureofaciens, dubbed "Chassis2.0," was developed by deleting endogenous gene clusters to minimize precursor competition. This chassis has shown high efficiency in producing diverse aromatic polyketides and serves as an excellent model for a versatile production platform [24].
Transformation Protocol
The following protocol details the transformation of a Streptomyces chassis, which is a critical step in establishing the biosynthetic machinery.
Materials:
- Streptomyces aureofaciens Chassis2.0 spores or mycelia [24]
- pET-28a-MoLAC14 plasmid DNA (or a suitable Streptomyces shuttle vector containing the gene of interest)
- Lysozyme solution (10 mg/mL)
- Sucrose solution (10.3%)
- Regeneration Medium (e.g., R2YE or R5 agar)
- Antibiotics for selection (e.g., apramycin)
Procedure:
- Preparation of Protoplasts: a. Inoculate a flask of TSB (Tryptic Soy Broth) with Streptomyces spores and incubate at 30°C with shaking until late-exponential growth phase (approx. 36-48 hours). b. Harvest the mycelia by centrifugation (3,000 - 4,000 × g for 10 minutes). c. Wash the pellet gently with 10.3% sucrose solution. d. Re-suspend the mycelia in a lysozyme solution (1-2 mg/mL in 10.3% sucrose) and incubate at 30°C for 30-60 minutes. Monitor protoplast formation under a microscope.
Transformation: a. Carefully pellet the protoplasts by gentle centrifugation (2,000 × g for 7-10 minutes). b. Wash the protoplast pellet twice with 10.3% sucrose to remove residual lysozyme. c. Re-suspend the protoplasts in a small volume of 10.3% sucrose. d. Aliquot protoplasts into microcentrifuge tubes and add the plasmid DNA (approx. 100-500 ng). Mix gently. e. Add an equal volume of 40% PEG 1000 (Polyethylene Glycol) to the protoplast-DNA mixture. Mix by gentle pipetting and incubate at room temperature for 1-2 minutes.
Regeneration and Selection: a. Dilute the transformation mixture with 1-2 mL of 10.3% sucrose. b. Plate the protoplasts onto Regeneration Medium (R2YE or R5 agar) supplemented with the appropriate antibiotic. c. Allow the plates to dry and incubate at 30°C for 5-7 days until transformant colonies appear.
Verification: a. Screen the resulting colonies by PCR using gene-specific primers for MoLAC14 to confirm the successful integration of the construct [5]. b. For strains harboring the complete biosynthetic pathway, confirm magnolol production via High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS) [5].
Flowchart of Streptomyces Transformation and Validation
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Magnolol Biosynthesis Research
| Reagent / Material | Function / Application | Example / Note |
|---|---|---|
| pET-28a Vector | Prokaryotic expression vector for gene cloning and protein expression in E. coli. | Contains T7 lac promoter for inducible expression [5]. |
| Golden Gate Assembly (GGA) | A modular cloning system for efficient and accurate assembly of multiple DNA fragments. | Enables 100% efficient construction of BGCs and mutant libraries [23]. |
| Gibson Assembly | An isothermal, single-reaction method for assembling multiple overlapping DNA fragments. | Used for constructing expression plasmids and BGCs [5]. |
| Chassis2.0 | An engineered Streptomyces aureofaciens strain optimized for production of aromatic polyketides. | Deletion of native BGCs reduces precursor competition [24]. |
| Chavicol | The direct precursor substrate for the final enzymatic step in magnolol synthesis. | Used in in vitro assays and as a fed substrate in fermentations [5]. |
| R2YE / R5 Agar | Regeneration media used for the recovery and growth of Streptomyces protoplasts after transformation. | Essential for obtaining transformants post-PEG treatment. |
| 7-Hydroxymethyl-10,11-MDCPT | 7-Hydroxymethyl-10,11-MDCPT, MF:C22H18N2O7, MW:422.4 g/mol | Chemical Reagent |
| Tricyclodecenyl acetate-13C2 | Tricyclodecenyl acetate-13C2, MF:C12H16O2, MW:194.24 g/mol | Chemical Reagent |
Analytical Methods for Validation
Confirming successful magnolol production is a multi-step process:
- HPLC (High-Performance Liquid Chromatography): Separates and analyzes the components of the culture extract, allowing for the identification of magnolol based on its retention time compared to an authentic standard.
- MS (Mass Spectrometry): Provides the molecular mass and fragmentation pattern of the produced compound, offering definitive confirmation of magnolol identity [5].
- GNPS Molecular Networking: This advanced technique can be used to identify not only the target compound but also related molecules and shunt products, which is particularly useful when refactoring pathways or generating mutant libraries [23].
Concluding Remarks
The construction of efficient biosynthetic machinery for magnolol production hinges on the synergistic application of optimized vector design, precise enzyme engineering, and the selection of a compatible microbial chassis. The protocols outlined here—from cloning the key laccase gene MoLAC14 and improving its properties through site-directed mutagenesis, to transforming a high-performance Streptomyces chassis—provide a robust framework for researchers. This approach, firmly situated within the principles of synthetic biology, enables the sustainable and scalable production of magnolol, facilitating further pharmacological studies and drug development efforts for this promising natural compound.
HERE IS THE DRAFT OF YOUR APPLICATION NOTES AND PROTOCOLS
Heterologous Expression of Key Enzymes: Case Study on MoLAC14 Production
Within the broader framework of developing a robust synthetic biology platform for the production of magnolol, a plant-derived compound with significant antimicrobial and therapeutic potential, the heterologous expression of key biosynthetic enzymes is a critical step [5]. The traditional extraction of magnolol from the bark of Magnolia officinalis is constrained by the plant's long growth cycle and the low concentration of the compound (~1%) [5] [8]. Furthermore, chemical synthesis routes often suffer from poor specificity, low yield, and environmental concerns [5] [8]. Synthetic biology offers a promising alternative, but its application for magnolol production has been hampered by a lack of understanding of its biosynthetic pathway in the plant [5].
Recent research has identified MoLAC14, a laccase enzyme from Magnolia officinalis, as a pivotal catalyst in the biosynthesis of magnolol, directly converting the precursor chavicol into magnolol [5]. This discovery provides a foundational genetic component for engineering a microbial cell factory. The functional expression of MoLAC14 in a heterologous host is therefore a cornerstone for enabling the sustainable bioproduction of magnolol. This document details the application notes and protocols for the heterologous production and engineering of MoLAC14, serving as a case study for the expression of key plant-derived enzymes in a synthetic biology context.
Key Findings and Quantitative Data
The identification and subsequent engineering of MoLAC14 have yielded critical quantitative data that inform the strategy for its heterologous production. The functional characterization confirmed its central role in magnolol biosynthesis, while protein engineering significantly enhanced its properties.
Table 1: Summary of Key Experimental Findings for MoLAC14
| Aspect | Finding | Implication for Heterologous Production |
|---|---|---|
| Catalytic Function | Confirmed to catalyze the one-step conversion of chavicol to magnolol [5]. | Validates MoLAC14 as the key gene for a simplified magnolol biosynthetic pathway in a heterologous host. |
| Critical Residue | Alanine scanning identified L532 as an essential residue; the L532A mutation enhanced activity [5]. | Indicates a key target for further rational protein engineering to improve enzyme performance. |
| Production Titre | The L532A variant boosted magnolol production to a level of 148.83 mg/L in vitro [5]. | Provides a benchmark for evaluating the success of heterologous expression and fermentation processes. |
| Thermal Stability | Mutations E345P, G377P, H347F, E346C, and E346F notably improved thermal stability [5]. | Crucial for industrial application; stable enzymes can withstand prolonged fermentation conditions and storage. |
Experimental Protocols
Gene Identification and Cloning of MoLAC14
The initial discovery of MoLAC14 involved comprehensive transcriptome sequencing of various M. officinalis tissues, which led to the identification of 30 potential laccase genes [5]. MoLAC14 was selected for functional characterization based on its high expression in magnolol-producing tissues and its presence in a putative biosynthetic gene cluster [5].
Protocol: Gene Cloning into a Prokaryotic Expression Vector
- Gene Synthesis: The coding sequence (CDS) of MoLAC14 was retrieved from the M. officinalis transcriptome and genome data. The gene was synthesized de novo with codon optimization for the intended expression host (e.g., E. coli) to improve translation efficiency [5].
- Vector Preparation: A pET-28a vector is linearized using restriction enzymes Nde I and Xho I [5]. This vector provides a T7 lac promoter for strong, inducible expression and an N-terminal hexahistidine (His6)-tag for simplified purification.
- Assembly and Transformation: The synthesized MoLAC14 gene is integrated into the linearized pET-28a vector using Gibson assembly [5]. The resulting recombinant plasmid is then transformed into an E. coli cloning strain, such as DH5α, for amplification. Transformants are selected on agar plates containing kanamycin.
- Plasmid Verification: Positive clones are cultured, and the plasmid DNA is isolated. The integrity of the construct, pET-28a-MoLAC14, is verified by analytical restriction digest and DNA sequencing.
Heterologous Expression inE. coli
This protocol outlines a standard procedure for expressing MoLAC14 in E. coli, a common host for initial protein production and characterization [25]. The following workflow visualizes the key stages from gene to purified protein.
Protocol: Expression and Purification
- Host Strain Transformation: The verified pET-28a-MoLAC14 plasmid is transformed into an E. coli expression host such as BL21(DE3). This strain is deficient in lon and ompT proteases to minimize protein degradation and contains the DE3 lysogen for T7 RNA polymerase expression [25].
- Culture and Induction:
- Inoculate a starter culture of LB medium supplemented with kanamycin and grow overnight at 37°C with shaking.
- Dilute the starter culture into fresh, baffled shaker flasks containing the same medium. Grow at 37°C with vigorous shaking (200-250 rpm) until the OD600 reaches 0.6-0.9.
- Lower the incubation temperature to 18°C to enhance proper protein folding and reduce inclusion body formation [25].
- Induce protein expression by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1-0.5 mM. Continue incubation with shaking overnight (~16-20 hours) [25].
- Cell Harvest and Lysis:
- Harvest the cells by centrifugation (e.g., 4,000 x g for 20 minutes at 4°C).
- Resuspend the cell pellet in a suitable lysis buffer (e.g., 50 mM Tris-HCl, 300 mM NaCl, 10 mM imidazole, pH 8.0) and lyse the cells using sonication or a homogenizer.
- Clarify the lysate by centrifugation at high speed (e.g., 15,000 x g for 30 minutes at 4°C) to remove cell debris.
- Protein Purification:
- Pass the clarified lysate through a chromatography column containing Ni-NTA resin, which binds the His6-tag on MoLAC14.
- Wash the resin extensively with wash buffer (e.g., 50 mM Tris-HCl, 300 mM NaCl, 20-50 mM imidazole, pH 8.0) to remove weakly bound contaminants.
- Elute the purified MoLAC14 protein using elution buffer containing a high concentration of imidazole (e.g., 250-500 mM).
- The purified protein can be desalted into an appropriate storage buffer (e.g., 50 mM Tris-HCl, pH 7.5, 10% glycerol) using dialysis or size-exclusion chromatography.
In Vitro Enzymatic Assay
To confirm the catalytic function of heterologously produced MoLAC14, an in vitro assay using its substrate, chavicol, is performed.
- Reaction Setup: Prepare a reaction mixture containing a suitable buffer (e.g., phosphate or citrate buffer, pH 5-6, optimal for laccase activity), the purified MoLAC14 enzyme, and chavicol as the substrate.
- Incubation: Incubate the reaction mixture at a defined temperature (e.g., 30°C) for a set period.
- Reaction Termination: Stop the reaction by denaturing the enzyme (e.g., by heat or acid addition) or by diluting the mixture with the mobile phase used for analysis.
- Product Analysis: Analyze the reaction products using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS) to detect and quantify the formation of magnolol, confirming the successful conversion of chavicol [5].
The Scientist's Toolkit: Research Reagent Solutions
The successful heterologous production and engineering of MoLAC14 rely on a suite of essential molecular biology and protein biochemistry reagents.
Table 2: Essential Research Reagents for MoLAC14 Production and Analysis
| Reagent / Tool | Function / Application | Example from MoLAC14 Study |
|---|---|---|
| pET-28a Vector | Prokaryotic expression vector; provides T7 promoter for high-level induction, His-tag for purification, and kanamycin resistance [5] [25]. | Used as the backbone for cloning and expressing the MoLAC14 gene in E. coli [5]. |
| BL21(DE3) E. coli | A common heterologous host for protein expression; deficient in proteases to enhance protein stability [25]. | Serves as the production host for recombinant MoLAC14 protein. |
| Ni-NTA Chromatography | Affinity purification technique that leverages the interaction between a His6-tag and immobilized nickel ions to purify recombinant proteins [25]. | Key step for purifying MoLAC14 after cell lysis. |
| Site-Directed Mutagenesis | A molecular technique to introduce specific point mutations into a gene sequence, enabling structure-function studies and protein engineering. | Used to generate stability-enhancing (e.g., E345P) and activity-boosting (L532A) mutants of MoLAC14 [5]. |
| HPLC & Mass Spectrometry | Analytical techniques for separating, detecting, and identifying chemical compounds; used to verify enzyme activity and product formation. | Essential for confirming the conversion of chavicol to magnolol by MoLAC14 in enzymatic assays [5]. |
| Ac-rC Phosphoramidite-15N | Ac-rC Phosphoramidite-15N, MF:C47H64N5O9PSi, MW:903.1 g/mol | Chemical Reagent |
| Enerisant hydrochloride | Enerisant hydrochloride, CAS:1152749-07-9, MF:C22H31ClN4O3, MW:435.0 g/mol | Chemical Reagent |
Pathway Engineering and Host Systems
Integrating the heterologously expressed MoLAC14 into a engineered microbial host is the ultimate goal for de novo magnolol production. This requires the reconstruction of the entire biosynthetic pathway. The diagram below illustrates the proposed complete synthetic biology pathway for magnolol production in a microbial host like yeast, starting from the central metabolite tyrosine.
While E. coli is a valuable host for initial protein production, other microbial systems offer distinct advantages for complex pathway engineering. The yeast Komagataella phaffii is a particularly robust platform, offering benefits such as high cell-density fermentation, strong inducible promoters (e.g., AOX1), and the ability to perform some post-translational modifications [26]. Optimization of physicochemical parameters like temperature, pH, and methanol concentration (for AOX1 induction) in K. phaffii can boost the yield of recombinant proteins by over 30% compared to baseline conditions [26]. Furthermore, other yeast strains like Ogataea minuta have been genetically engineered as production hosts by disrupting genes like AOX1 (to control metabolism) and PRB1 (a protease to reduce protein degradation), further enhancing their capability for industrial-scale manufacturing of heterologous proteins [27]. The choice of host system depends on the specific requirements of the pathway, including the need for compartmentalization, the type of post-translational modifications required by the enzymes, and the scalability of the process.
Efficient protein production is a cornerstone of synthetic biology, enabling the manufacturing of therapeutic compounds, enzymes, and industrial biocatalysts. Within the context of magnolol production—a valuable natural compound with demonstrated antibacterial, anti-inflammatory, and anti-cancer properties—optimizing protein production becomes essential for developing viable biosynthetic pathways [28] [8]. Traditional extraction of magnolol from Magnolia officinalis bark requires 10-15 years of plant growth and yields low concentrations approximately 1%, making it economically and environmentally challenging [5] [10]. Synthetic biology approaches that harness engineered proteins offer a promising alternative to conventional production methods, potentially enabling sustainable, high-yield manufacturing of this valuable compound and similar natural products.
This Application Note presents strategic frameworks and detailed protocols for optimizing protein production systems, with specific applications to magnolol biosynthesis. We integrate computational design, strain engineering, and cell-free systems to address key bottlenecks in protein expression, stability, and functionality, providing researchers with practical tools to accelerate their bioproduction pipelines.
Strategic Framework for Protein Production Optimization
AI-Driven Platform and Strain Engineering
Automated DBTL Pipeline: Implementing a fully automated Design-Build-Test-Learn (DBTL) cycle dramatically accelerates protein optimization. Recent advances demonstrate that ChatGPT-4 can generate experimental design code without manual revision, significantly reducing development time [29]. This approach integrates active learning with a cluster margin strategy that selects both informative and diverse experimental conditions, maximizing learning efficiency while minimizing experimental iterations. When applied to antimicrobial protein production, this framework achieved 2- to 9-fold yield improvements in just four cycles [29].
Strain Engineering for Magnolol Precursors: For magnolol biosynthesis, engineering microbial factories requires optimizing multiple enzymatic steps. The proposed biosynthetic pathway begins with tyrosine conversion to p-coumaryl alcohol through enzymes including tyrosine ammonia-lyase (TAL), 4-coumarate CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), and alcohol dehydrogenase (ADH) [5] [10]. Subsequent steps involve coniferyl alcohol acetyltransferase (CAAT) and allylphenol synthases (APS) to produce chavicol, with final conversion to magnolol catalyzed by laccase enzymes [5]. Prioritizing expression optimization of these key enzymes, particularly the rate-limiting steps, is essential for efficient magnolol production.
Table 1: Key Enzymes for Magnolol Biosynthetic Pathway
| Enzyme | Function in Pathway | Engineering Approach |
|---|---|---|
| Tyrosine ammonia-lyase (TAL) | Converts tyrosine to p-coumaric acid | Codon optimization, promoter engineering |
| 4-coumarate CoA ligase (4CL) | Activates p-coumaric acid to CoA ester | Protein fusion strategies |
| Cinnamoyl-CoA reductase (CCR) | Reduces feruloyl-CoA to coniferaldehyde | N-terminal truncation for solubility |
| Alcohol dehydrogenase (ADH) | Converts coniferaldehyde to coniferyl alcohol | Cofactor balancing |
| Laccase (MoLAC14) | Oxidatively couples chavicol to magnolol | Thermal stability engineering |
Cell-Free Systems and Enzyme Engineering
Cell-Free Protein Synthesis (CFPS): CFPS platforms bypass cellular viability constraints, enabling rapid prototyping of protein production systems. These minimal systems contain only essential components for transcription and translation, allowing precise control over reaction conditions [29]. For magnolol biosynthesis, CFPS is particularly valuable for screening laccase variants without the constraints of cellular metabolism or potential toxicity. The modular nature of CFPS allows systematic optimization of magnesium concentration, energy sources, and redox components to enhance protein yields.
Enzyme Engineering for Enhanced Function: Protein engineering plays a crucial role in optimizing key enzymes for magnolol production. Research has identified MoLAC14 as a pivotal laccase enzyme responsible for magnolol synthesis through oxidative coupling of chavicol [5] [10]. Targeted mutations to enhance thermal stability (E345P, G377P, H347F, E346C, E346F) significantly improved enzyme performance, while the L532A mutation boosted magnolol production to 148.83 mg/L—an unprecedented yield demonstrating the power of precise enzyme engineering [5]. Alanine scanning further identified essential residues, providing a roadmap for future engineering efforts.
Experimental Protocols
Protocol 1: Automated DBTL for Protein Production Optimization
Application: This protocol is adapted from the AI-driven workflow that achieved 2- to 9-fold yield improvements for colicin proteins and can be applied to optimize magnolol biosynthetic enzymes [29].
Materials:
- Liquid handling robot
- Microplates
- Cell-free protein synthesis system
- E. coli or HeLa cell extracts
- DNA templates encoding target proteins
- Fluorescence detection system
Procedure:
- Design Phase: Use ChatGPT-4 or similar LLM to generate Python scripts for experimental design without manual revision.
- Build Phase: Automate reaction assembly using liquid handlers to dispense CFPS components with varying compositions.
- Test Phase: Incubate reactions at 30°C for 4-6 hours, then quantify protein yields using fluorescence measurements.
- Learn Phase: Apply active learning with cluster margin sampling to select the most informative subsequent experiments.
- Iteration: Repeat cycles 3-5 times, with each cycle informing the next experimental design.
Technical Notes: The key advantage of this approach is the active learning strategy that selects experiments balancing diversity and uncertainty, maximizing information gain per experimental cycle. For magnolol pathway enzymes, focus optimization on magnesium concentration (1-10 mM), energy regeneration system components, and expression temperature (25-37°C).
Protocol 2: Engineering Thermal Stability in Laccase Enzymes
Application: This protocol details the enzyme engineering approach that enhanced MoLAC14 stability and magnolol production [5].
Materials:
- pET-28a expression vector
- BL21(DE3) E. coli strain
- Site-directed mutagenesis kit
- Chavicol substrate
- Copper (II) ions (Cu²âº)
- HPLC system with mass spectrometry
Procedure:
- Gene Synthesis: Synthesize MoLAC14 CDS with codon optimization for E. coli expression.
- Vector Construction: Clone into pET-28a between NdeI and XhoI sites using Gibson assembly.
- Mutagenesis: Introduce stability-enhancing mutations (E345P, G377P, H347F, E346C, E346F) via site-directed mutagenesis.
- Protein Expression: Culture E. coli at 37°C, induce with 0.5 mM IPTG, and add 0.5 mM Cu²⺠to culture.
- Enzyme Assay: Incubate purified enzyme variants with 2 mM chavicol in phosphate buffer (pH 7.0) at 30°C for 2 hours.
- Product Analysis: Quantify magnolol production using HPLC-MS.
Technical Notes: The L532A mutation proved particularly effective for enhancing magnolol production. Include this mutation in screening efforts. Monitor enzyme half-life at 40°C to assess thermal stability improvements.
Protocol 3: Integrated Fermentation for Magnolol Production
Application: This protocol enables coproduction of magnolol and related compounds from herbal extraction residues, maximizing resource utilization [30].
Materials:
- Magnolia officinalis residues (MOR)
- NaOH solution
- MIL-101(Cr) metal-organic framework
- Engineered yeast strain for β-amyrin production
- Cellulolytic enzymes
Procedure:
- Pretreatment: Treat MOR with 2% NaOH at 80°C for 2 hours to release bound magnolol and honokiol.
- Adsorption: Apply MIL-101(Cr) to absorb magnolol and honokiol from pretreated solution.
- Enzymatic Saccharification: Incubate pretreated MOR with cellulases at 50°C for 48 hours.
- Fermentation: Inoculate MOR hydrolysate with engineered yeast for β-amyrin production.
- Extraction: Recover magnolol, honokiol, and β-amyrin using organic solvents.
Technical Notes: This integrated approach yields approximately 1 kg honokiol, 8 kg magnolol, and 7.64 kg β-amyrin per ton of MOR, with a total economic output of approximately 170,700 RMB [30].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for Optimizing Magnolol Biosynthesis
| Reagent | Function | Application Notes |
|---|---|---|
| pET-28a vector | Protein expression | Contains T7 promoter for high-level expression in E. coli [5] |
| MIL-101(Cr) | Metal-organic framework | Maximum absorption capacity of 255.64 mg/g for magnolol and honokiol [30] |
| Chavicol | Laccase substrate | Starting material for magnolol production via oxidative coupling [5] |
| Copper (II) ions | Cofactor | Essential for laccase activity; add to culture at 0.5 mM concentration [5] |
| Engineered yeast | β-amyrin production | Enables coproduction from MOR hydrolysates; yield of 382 mg/L [30] |
| 3-Ethyl-5-methyl-2-vinylpyrazine-d3 | 3-Ethyl-5-methyl-2-vinylpyrazine-d3, MF:C9H12N2, MW:151.22 g/mol | Chemical Reagent |
| E3 Ligase Ligand-linker Conjugate 12 | E3 Ligase Ligand-linker Conjugate 12, MF:C30H39N5O6, MW:565.7 g/mol | Chemical Reagent |
Workflow Visualization
AI-Driven Protein Optimization Workflow
Magnolol Biosynthetic Pathway
The integration of AI-driven optimization, strategic enzyme engineering, and advanced fermentation platforms provides a powerful framework for enhancing protein production in synthetic biology applications. For magnolol biosynthesis specifically, these approaches have demonstrated remarkable success, with engineered laccase variants achieving unprecedented production levels. Future developments will likely focus on further integration of multi-omics data, de novo protein design, and expanded cell-free platforms to accelerate the design of efficient biosynthetic pathways. As these technologies mature, they will undoubtedly facilitate the transition from laboratory-scale protein production to industrial-scale manufacturing of magnolol and other valuable natural compounds.
Pathway Assembly and Metabolic Engineering for Precursor Supplementation
Synthetic biology offers a promising avenue for the sustainable production of plant-derived natural products like magnolol, a hydroxylated biphenyl compound from Magnolia officinalis with demonstrated anticancer, antimicrobial, and anti-inflammatory properties [28]. Traditional extraction from magnolia bark is constrained by the plant's long cultivation time (10-15 years) and low magnolol concentration (approximately 1%) [5]. Chemical synthesis, while achievable, often suffers from low yield, formation of by-products, and significant environmental pollution due to high organic solvent consumption [5]. Pathway assembly and metabolic engineering in microbial hosts present a viable alternative to overcome these limitations, enabling efficient, scalable, and environmentally friendly magnolol production. This Application Note details the biosynthetic pathway elucidation and provides protocols for the metabolic engineering of microbial chassis to supplement key precursors for magnolol biosynthesis.
Biosynthetic Pathway Identification and Validation
Proposed Magnolol Biosynthetic Pathway
Recent research has proposed a biosynthetic pathway for magnolol originating from primary metabolism. The pathway is hypothesized to commence with tyrosine, leading to the key intermediate chavicol, which is then dimerized to form magnolol [5]. The table below outlines the key enzymatic steps and precursors.
Table 1: Proposed Biosynthetic Pathway from Tyrosine to Magnolol
| Precursor/Intermediate | Enzyme | Reaction / Role |
|---|---|---|
| L-Tyrosine | Tyrosine ammonia-lyase (TAL) | Deamination to yield coumaric acid |
| p-Coumaric acid | 4-Coumarate CoA ligase (4CL) | Activation to coumaroyl-CoA |
| p-Coumaryl-CoA | Cinnamoyl-CoA reductase (CCR) | Reduction to cinnamyl aldehyde |
| Coniferyl alcohol / p-Coumaryl alcohol | Alcohol Dehydrogenase (ADH) | Reduction to the corresponding alcohol |
| Chavicol | Allylphenol Synthases (APS) | Formation of the allylphenol scaffold |
| Magnolol | Laccase (e.g., MoLAC14) | Oxidative coupling of two chavicol molecules |
Key Enzyme: MoLAC14 Laccase
The conversion of chavicol to magnolol is a one-step oxidative coupling reaction catalyzed by laccase [5]. From 30 potential laccase genes identified in M. officinalis, MoLAC14 was functionally validated as the pivotal enzyme in magnolol synthesis [5].
Table 2: Key Enzymatic Activity for Magnolol Production
| Enzyme | Function in Pathway | Experimental Validation | Production Titer |
|---|---|---|---|
| MoLAC14 | Oxidative dimerization of chavicol to magnolol | In vitro assay with chavicol substrate; product confirmed via HPLC and MS [5]. | 148.83 mg/L (with L532A mutation) [5] |
Engineering of MoLAC14 through site-directed mutagenesis has led to improved performance. Mutations such as E345P, G377P, H347F, E346C, and E346F enhanced thermal stability, while the L532A mutation significantly boosted magnolol production to 148.83 mg/L in vitro, demonstrating the potential of enzyme engineering for yield improvement [5].
Metabolic Engineering Strategies for Precursor Supplementation
Successful heterologous production of magnolol requires engineering the host's metabolic network to ensure abundant supply of key precursors, primarily L-tyrosine and chavicol. The following strategies and protocols can be implemented.
Engineering the Shikimate Pathway for L-Tyrosine Overproduction
The shikimate pathway is the primary source of aromatic amino acids. To enhance carbon flux towards L-tyrosine:
- Overexpress Key Enzymes: Amplify the expression of rate-limiting enzymes in the shikimate pathway, such as 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (AroG) and chorismate mutase/prephenate dehydrogenase (TyrA). Use feedback-resistant mutants (e.g., AroGfbr) to circumvent endogenous regulation [31].
- Downcompete Branch Pathways: Attenuate or knockout genes directing flux towards competing amino acids phenylalanine and tryptophan (e.g., pheA, trpED).
- Enhance Cofactor Regeneration: Ensure sufficient supply of cofactors like NADPH, which is required by TyrA.
Protocol: Modular Engineering of the Shikimate Pathway
Objective: To construct a microbial strain with enhanced L-tyrosine production. Principle: This protocol employs Multivariate Modular Metabolic Engineering (MMME) to balance the expression of genes in the shikimate pathway [31]. The pathway is divided into two modules to optimize flux independently.
Materials:
- Plasmids: A library of expression vectors with varying promoter strengths (e.g., J23100, J23101, J23104 from the Anderson promoter library) and RBS sequences [32].
- Strain: An E. coli or S. cerevisiae production chassis with deletions in competitive pathways (ΔpheA, ΔtrpED).
- Reagents: PCR reagents, Gibson assembly or Golden Gate assembly mix, transformation-competent cells, LB or appropriate selective media, antibiotics.
Procedure:
- Pathway Segmentation:
- Module 1 (Core Shikimate): Contains genes arogfbr, arob, aroc, arod, aroe.
- Module 2 (L-Tyrosine Branch): Contains genes arol, tyrafbr.
Combinatorial Assembly:
- Assemble multiple variants of each module by using different promoters and RBSs for each gene. This creates a library of genetic constructs for each module.
Screening for Optimal Producers:
- Co-transform the host strain with combinations of Module 1 and Module 2 variants.
- Screen the resulting strain library for L-tyrosine production using HPLC or colorimetric assays.
- Select the top-performing strain for subsequent integration of the chavicol and magnolol synthesis modules.
Protocol: Heterologous Expression of the Chavicol-to-Magnolol Module
Objective: To introduce and optimize the final steps of magnolol biosynthesis in a microbial host. Principle: The genes for the chavicol biosynthesis (APS) and dimerization (MoLAC14) are introduced into the L-tyrosine-overproducing strain.
Materials:
- Gene Constructs: Codon-optimized genes for APS and MoLAC14 (or its engineered variant, e.g., L532A) cloned into an expression vector.
- Strain: High L-tyrosine producing strain from Protocol 3.2.
- Analytical Equipment: HPLC-MS for magnolol detection and quantification.
Procedure:
- Vector Construction:
- Clone the APS and MoLAC14 genes into a compatible expression plasmid(s) under the control of inducible promoters (e.g., pBAD, T7, or LacO1 [32]).
Strain Transformation:
- Introduce the constructed plasmid(s) into the engineered L-tyrosine production host.
Fermentation and Induction:
- Inoculate a shake flask with the transformed strain and grow in selective medium to mid-exponential phase.
- Induce gene expression with the appropriate inducer (e.g., IPTG for LacO1).
- Supplement the culture with copper ions (Cu2+), as laccases are copper-dependent enzymes.
Product Analysis:
- After 24-48 hours of post-induction incubation, extract metabolites from the culture broth.
- Analyze the extract using HPLC-MS. Magnolol can be identified by comparing its retention time and mass spectrum with an authentic standard (m/z 265 [M+H]+).
The Scientist's Toolkit: Research Reagent Solutions
The table below lists essential materials and reagents required for the metabolic engineering and production of magnolol.
Table 3: Key Research Reagents for Magnolol Pathway Engineering
| Reagent / Tool | Function / Application | Examples / Specifications |
|---|---|---|
| Chassis Strains | Host for heterologous pathway expression | Escherichia coli BL21(DE3), Saccharomyces cerevisiae |
| Expression Vectors | Cloning and expression of pathway genes | pET-28a (for MoLAC14) [5], pBb series, pRSFDuet |
| Regulatory Elements | Tunable control of gene expression | Promoters (LacO1, Tac, pBAD), RBSs (AGGAGA/T consensus) [32] |
| Gene Editing System | Precise genomic modifications | CRISPR/Cas9 system for K. oryzendophytica [32] |
| Key Heterologous Genes | Enzymes for the target pathway | MoLAC14 (laccase), APS (allylphenol synthase) |
| Analytical Standards | Product identification and quantification | Magnolol (purity ≥ 98%) [33], Honokiol |
| Analytical Equipment | Detection and quantification of metabolites | HPLC-MS, GC-MS |
| 2-Isopropyl-5-methylpyrazine-d3 | 2-Isopropyl-5-methylpyrazine-d3, MF:C8H12N2, MW:139.21 g/mol | Chemical Reagent |
| KRAS G12D modulator-1 | KRAS G12D modulator-1, MF:C30H36FN5O4, MW:549.6 g/mol | Chemical Reagent |
Pathway and Engineering Workflow Diagrams
The following diagrams illustrate the complete magnolol biosynthetic pathway and the overall metabolic engineering workflow.
Diagram 1: Magnolol biosynthetic pathway from glucose. Key engineered precursors (L-Tyrosine, Chavicol) are highlighted. The critical dimerization step catalyzed by MoLAC14 is marked in red.
Diagram 2: Metabolic engineering workflow for microbial magnolol production, highlighting critical stages for precursor supplementation and pathway optimization.
Overcoming Production Hurdles: Enzyme Engineering and Process Optimization
The efficient production of valuable compounds like magnolol, a natural product with demonstrated antibacterial properties, using synthetic biology is often hampered by the inherent limitations of native enzymes [10]. These biocatalysts frequently exhibit insufficient activity, stability, or compatibility with industrial process conditions, leading to low yields and impractical production economics [34] [35]. To overcome these hurdles, enzyme engineering has emerged as a critical discipline, with rational design and directed evolution representing two powerful, complementary strategies [36]. This protocol details the application of these methods to enhance enzyme performance, with specific examples drawn from the context of optimizing laccase enzymes for the synthesis of magnolol from chavicol [10].
Core Principles and Comparison of Strategies
Rational design relies on a deep understanding of the relationship between an enzyme's structure and its function. Using computational models and structural data, specific amino acid residues are targeted for mutation to precisely alter enzyme properties [37] [38]. In contrast, directed evolution mimics natural selection in a laboratory setting. It involves the generation of vast libraries of random enzyme variants followed by high-throughput screening or selection to identify individuals with improved traits, without requiring prior structural knowledge [36] [39].
The table below summarizes the key characteristics of each approach.
Table 1: Comparison of Rational Design and Directed Evolution
| Feature | Rational Design | Directed Evolution |
|---|---|---|
| Prerequisite Knowledge | Requires detailed structural and mechanistic information (e.g., from crystal structures, homology models, or advanced computational tools like ABACUS-T) [37] [38]. | Requires no prior structural knowledge; only a reliable screening or selection assay is needed [39]. |
| Methodology | Targeted, site-specific mutagenesis of predetermined residues. | Random mutagenesis of the entire gene or focused regions. |
| Library Size | Small and focused, containing dozens to hundreds of variants [38]. | Very large, containing thousands to millions of variants [39]. |
| Primary Challenge | Accurate prediction of mutation effects; risk of functional loss if critical residues are altered [37]. | Development of a robust, high-throughput screening method; can be resource-intensive [36] [39]. |
| Typical Outcome | Fewer, highly optimized mutations; can introduce dramatic stability enhancements (e.g., ∆Tm ≥ 10°C) with a handful of designs [38]. | Incremental improvements accumulated over multiple evolution cycles. |
Experimental Protocols
Rational Design for Enhanced Thermostability
This protocol uses the engineering of a laccase (MoLAC14) for improved magnolol production as a case study [10].
1. Identify Target Residues:
- Structural Analysis: Use a crystal structure or a high-confidence predicted model (e.g., from AlphaFold or ABACUS-T) of the enzyme to identify flexible regions or loops critical for stability [38].
- Computational Redesign: Employ an inverse folding model like ABACUS-T. Input the enzyme's backbone structure, optionally with a bound ligand (e.g., chavicol) and multiple sequence alignment (MSA) data to preserve functional residues. The model will output a redesigned sequence with dozens of mutations aimed at optimizing stability while maintaining function [38].
2. Library Design and Generation:
- Site-Directed Mutagenesis: For a focused approach, design primers to introduce specific point mutations (e.g., E345P, G377P in MoLAC14) suspected to improve stability by reducing backbone flexibility or introducing favorable interactions [10].
- Gene Synthesis: For computationally designed sequences from tools like ABACUS-T, the entire optimized gene sequence is synthesized de novo [38].
3. Expression and Purification:
- Clone the wild-type and mutant genes into an appropriate expression vector (e.g., pET-28a).
- Chemically transform the plasmid into a suitable expression host, such as E. coli BL21(DE3).
- Culture the cells, induce protein expression with IPTG, and supplement the medium with necessary cofactors (e.g., Cu²⺠for laccases).
- Purify the enzymes using affinity chromatography for functional characterization [10].
4. Screening for Thermostability:
- Melting Temperature (Tm) Assay: Use differential scanning fluorimetry (DSF) to determine the Tm of the enzyme variants. A significant increase in Tm (e.g., ∆Tm ≥ 10°C) indicates improved thermostability [38].
- Residual Activity Assay: Incubate the wild-type and mutant enzymes at an elevated temperature (e.g., 40-60°C) for a set duration. Periodically withdraw samples, cool them on ice, and measure the residual activity under standard assay conditions. The mutant with higher residual activity after heating is more stable [35] [10].
5. Functional Validation:
- Assess the catalytic activity of the stabilized mutants to ensure functionality is not compromised. For MoLAC14, this involves testing its ability to convert chavicol to magnolol in vitro, followed by quantification using HPLC and MS [10].
- Determine kinetic parameters (Km, kcat) to evaluate any changes in catalytic efficiency.
Directed Evolution for Enhanced Activity
This protocol outlines a general directed evolution campaign, applicable for improving traits like enzyme activity or solvent tolerance [36] [39].
1. Diversity Generation:
- Random Mutagenesis: Use error-prone PCR (epPCR) or mutator strains to introduce random mutations across the entire gene. This creates a large library of enzyme variants.
2. Library Screening/Selection:
- High-Throughput Screening (HTS): Develop a microtiter plate-based assay to rapidly screen thousands of variants. For oxidases like laccases, this could be a colorimetric assay where product formation generates a detectable signal.
- Selection Systems: If possible, engineer a selection system where enzyme activity is coupled to host cell survival, dramatically increasing screening throughput [39].
3. Iterative Rounds of Evolution:
- Isolate the top-performing variants from the first screening round.
- Use these improved variants as templates for the next round of mutagenesis and screening.
- Repeat this process iteratively until the desired level of performance is achieved. Accumulating beneficial mutations over multiple rounds can lead to significant improvements [36].
4. Characterization of Evolved Mutants:
- Sequence the evolved genes to identify the accumulated mutations.
- Express, purify, and thoroughly characterize the final evolved enzyme(s) for activity, stability, and specificity, as described in Section 3.1.
Key Research Reagent Solutions
The table below lists essential reagents and their applications for enzyme engineering experiments.
Table 2: Key Research Reagents and Materials
| Reagent / Material | Function / Application | Example & Notes |
|---|---|---|
| Expression Vector | Carries the gene of interest for protein expression in a host. | pET-28a vector for high-level expression in E. coli [10]. |
| Expression Host | Cellular system for producing the recombinant enzyme. | E. coli BL21(DE3) strain [10]. |
| Inducer | Triggers the expression of the target gene. | Isopropyl β-D-1-thiogalactopyranoside (IPTG) [10]. |
| Affinity Chromatography Resin | Purifies the enzyme based on a specific tag. | Ni-NTA resin for purifying His-tagged proteins [10]. |
| Restriction Enzymes | Used for cloning genes into expression vectors. | High-fidelity (Time-Saver) enzymes from NEB to prevent star activity [40]. |
| DNA Cleanup Kits | Purify DNA fragments (e.g., after PCR) to remove inhibitors. | Monarch PCR & DNA Cleanup Kit [40]. |
| Buffer Components | Maintain optimal pH and environment for enzyme reactions. | Use recommended buffers (e.g., NEBuffers) and note that some are now BSA-free, containing rAlbumin [40]. |
| Cofactors / Substrates | Essential for enzyme activity and screening assays. | Copper (II) ions (Cu²âº) for laccase activity; chavicol as substrate for magnolol production [10]. |
Data Presentation and Analysis
The following table quantifies the success of a rational design campaign for the laccase MoLAC14, demonstrating significant improvements in both stability and productivity for magnolol synthesis [10].
Table 3: Quantitative Results from Engineering MoLAC14 Laccase
| Enzyme Variant | Key Mutation(s) | Impact on Thermostability | Magnolol Production (mg/L) |
|---|---|---|---|
| Wild-type MoLAC14 | - | Baseline | Baseline (Unspecified) |
| Stabilized Mutant 1 | E345P | Improved | Improved vs. WT |
| Stabilized Mutant 2 | G377P | Improved | Improved vs. WT |
| Stabilized Mutant 3 | H347F | Improved | Improved vs. WT |
| Stabilized Mutant 4 | E346C | Improved | Improved vs. WT |
| Stabilized Mutant 5 | E346F | Improved | Improved vs. WT |
| High-Activity Mutant | L532A | Not Specified | 148.83 |
Concluding Remarks
The synergistic application of rational design and directed evolution provides a robust framework for overcoming the limitations of natural enzymes in synthetic biology pathways. As demonstrated in the engineering of laccases for magnolol production, rational design can yield highly stable and active catalysts [10]. The continued advancement of computational tools, such as the ABACUS-T model which integrates structural and evolutionary data, promises to further streamline the design process, enabling the creation of superior biocatalysts with fewer experimental iterations [38]. These engineered enzymes are pivotal for developing efficient, scalable, and economically viable bioprocesses for high-value compounds like magnolol.
In the burgeoning field of synthetic biology, the production of high-value plant-derived compounds like magnolol faces a significant hurdle: the instability of key biosynthetic enzymes under industrial process conditions. Magnolol, a potent antibacterial and anticancer compound extracted from Magnolia officinalis, has seen its synthetic biological production hampered by the limited thermal robustness of the enzymes involved in its pathway [10] [8] [28]. Recent breakthroughs have identified the laccase enzyme MoLAC14 as a pivotal catalyst for the one-step conversion of chavicol to magnolol [10] [5]. This application note details how strategic mutations, including E345P and G377P, significantly enhance the thermal stability of MoLAC14, thereby laying the groundwork for efficient, industrially viable magnolol production. We present quantitative stability data, detailed protocols for assessing thermostability, and essential tools for implementing these advances.
Key Findings: Mutation-Induced Thermal Stabilization of MoLAC14
Directed efforts to engineer MoLAC14 have identified several point mutations that confer improved thermal stability, a critical factor for enzymatic activity and longevity during bioprocessing. The following table summarizes the key mutations and their documented impact on the enzyme's stability profile [10].
Table 1: Impact of Key Mutations on MoLAC14 Thermal Stability
| Mutation | Reported Effect on Thermal Stability |
|---|---|
| E345P | Notable enhancement |
| G377P | Notable enhancement |
| H347F | Notable enhancement |
| E346C | Notable enhancement |
| E346F | Notable enhancement |
| L532A | Boosted magnolol production to 148.83 mg/L |
Beyond single-point mutations, comprehensive alanine scanning of MoLAC14 identified essential residues and revealed that the L532A mutation not only contributed to stability but also dramatically increased magnolol titers in a production setup, achieving an unprecedented yield of 148.83 mg/L [10]. This demonstrates that stability engineering can have a direct and profound impact on overall pathway efficiency and productivity.
Experimental Protocols for Thermostability Assessment
To validate the effects of these mutations, researchers must employ robust and reproducible assays. Below is a detailed protocol for a critical experiment cited in the foundational research.
Protocol: In Vitro Thermostability Assay via Residual Activity Measurement
Objective: To determine the half-life and residual activity of wild-type versus mutant MoLAC14 after exposure to elevated temperatures.
Principle: The enzyme is incubated at a target temperature for varying durations. The residual activity is measured by assessing its capability to convert chavicol to magnolol, which is quantified using High-Performance Liquid Chromatography (HPLC) [10].
Materials:
- Purified wild-type or mutant MoLAC14 enzyme
- Chavicol substrate
- Appropriate reaction buffer (e.g., phosphate buffer, pH 7.0)
- Thermostatic water bath or thermal cycler
- HPLC system with a UV/Vis or mass spectrometry (MS) detector
Procedure:
- Enzyme Incubation: Dilute the purified enzyme to a standardized concentration in a suitable buffer. Aliquot the enzyme solution into low-protein-binding microcentrifuge tubes.
- Heat Challenge: Place the tubes in a pre-heated thermostatic block set at the desired challenge temperature (e.g., 40°C, 45°C, 50°C). Remove tubes in triplicate at predetermined time intervals (e.g., 0, 5, 15, 30, 60 minutes) and immediately transfer them to an ice bath to halt thermal denaturation.
- Activity Assay: For each time point, initiate the enzymatic reaction by adding an equal volume of substrate solution containing chavicol to the cooled enzyme. Allow the reaction to proceed for a fixed period under optimal conditions (e.g., 30°C).
- Reaction Termination & Quantification: Stop the reaction by adding an equal volume of an organic solvent like methanol. Precipitated protein can be removed by centrifugation.
- Magnolol Quantification: Analyze the supernatant using HPLC. Separations can be achieved on a reversed-phase C18 column using a water-acetonitrile gradient. Magnolol is typically detected by its UV absorbance and confirmed by its specific mass-to-charge ratio (m/z) via MS. Quantify the magnolol peak area against a standard curve.
- Data Analysis: The residual activity at each time point is calculated as a percentage of the activity of the unheated control (0-minute time point). Plot the natural logarithm of residual activity versus time; the negative slope of the linear fit is the inactivation rate constant (k). The half-life (t_{1/2}) is then calculated as ln(2)/k.
Workflow for Engineering and Validating Thermostable Laccase Variants
The process of developing and characterizing stabilized enzyme variants like MoLAC14-E345P/G377P follows a logical, multi-stage workflow. The diagram below outlines the key steps from library creation to final validation.
Diagram Title: Thermostable Enzyme Engineering Workflow
The Scientist's Toolkit: Essential Research Reagents
To successfully replicate and build upon this research, scientists require a specific set of reagents and tools. The following table catalogues the essential components used in the identification and validation of stable MoLAC14 variants [10].
Table 2: Essential Research Reagents for MoLAC14 Engineering
| Reagent / Tool | Function in Research | Specific Example / Note |
|---|---|---|
| Expression Vector | Heterologous overexpression of the laccase gene. | pET-28a vector for expression in E. coli BL21(DE3). |
| Inducer | To induce the expression of the recombinant gene. | Isopropyl β-D-1-thiogalactopyranoside (IPTG). |
| Cofactor | Essential for the catalytic activity of many laccases. | Copper (II) ions (Cu²âº). |
| Chromatography System | Separation and quantification of magnolol from reaction mixtures. | High-Performance Liquid Chromatography (HPLC) system. |
| Detection Method | Confirmatory identification of the magnolol product. | Mass Spectrometry (MS), often coupled with HPLC. |
| Thermal Shift Assay | High-throughput method to determine protein melting temperature (Tm). | Differential Scanning Fluorimetry (DSF). |
The targeted introduction of mutations such as E345P and G377P represents a validated and powerful strategy for overcoming the thermal instability limitations of key enzymes in synthetic biology pathways. The application of the protocols and tools outlined herein will enable researchers to systematically engineer and characterize robust biocatalysts. For magnolol production, the successful stabilization of MoLAC14 marks a critical advancement, paving the way for sustainable and economically feasible biomanufacturing of this therapeutically important compound. Future work will likely focus on combining the most beneficial mutations and applying these principles to other fragile enzymes within the magnolol and related natural product biosynthetic pathways.
This application note provides a detailed protocol for using alanine scanning mutagenesis to identify and optimize key residues in enzymes for enhanced catalytic efficiency in synthetic biology pathways. We demonstrate the methodology through a case study on MoLAC14, a laccase enzyme critical for the synthetic production of magnolol, a plant-derived compound with significant pharmaceutical value. The L532A mutation in MoLAC14 successfully increased magnolol production to 148.83 mg/L, showcasing the power of this approach for metabolic engineering [10]. The following sections offer a complete experimental workflow, from gene construction to product quantification, enabling researchers to apply these techniques to other enzyme optimization projects.
Synthetic biology offers a promising route for the sustainable production of high-value natural compounds like magnolol, which possesses potent antibacterial, anti-inflammatory, and antioxidative properties [10] [9] [41]. However, the efficiency of biosynthetic pathways is often limited by the catalytic performance of key enzymes. Alanine scanning serves as a powerful technique for functional epitope mapping, allowing researchers to determine the contribution of specific amino acid residues to a protein's stability and function [42] [43].
By systematically replacing target residues with alanine, which features a non-bulky, chemically inert methyl side chain, researchers can probe the role of side chains without drastically altering the protein's secondary structure [42]. This protocol details the application of alanine scanning to identify residue L532 in the laccase enzyme MoLAC14, a mutation that profoundly enhanced magnolol synthesis, providing a blueprint for similar enzyme engineering endeavors [10].
Key Research Reagent Solutions
The table below lists essential reagents and materials required for the experiments described in this protocol.
Table 1: Essential Research Reagents and Materials
| Reagent/Material | Function/Application | Example Source / Reference |
|---|---|---|
| pET-28a Vector | Protein expression vector for cloning and expression in E. coli | [10] |
| E. coli BL21(DE3) | Host strain for recombinant protein expression | [10] |
| Chavicol | Substrate for the laccase-catalyzed reaction to produce magnolol | [10] |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Chemical inducer for protein expression in E. coli | [10] |
| Copper (II) ions (Cu²âº) | Cofactor for laccase enzyme activity | [10] |
| High-Performance Liquid Chromatography (HPLC) | System for quantifying magnolol production | [10] |
| Mass Spectrometry (MS) | System for confirming magnolol identity | [10] |
The following table summarizes the key mutations in MoLAC14 and their respective effects on enzyme performance and magnolol production, as identified through alanine scanning and other mutagenesis strategies [10].
Table 2: Impact of Key MoLAC14 Mutations on Enzyme Properties and Magnolol Yield
| Mutation | Effect on Thermal Stability | Effect on Catalytic Activity | Magnolol Production |
|---|---|---|---|
| E345P | Enhanced | Not Specified | Not Specified |
| G377P | Enhanced | Not Specified | Not Specified |
| H347F | Enhanced | Not Specified | Not Specified |
| E346C | Enhanced | Not Specified | Not Specified |
| E346F | Enhanced | Not Specified | Not Specified |
| L532A | Not Specified | Enhanced | 148.83 mg/L |
Experimental Protocol
Gene Synthesis and Plasmid Construction
This section outlines the steps for preparing the gene construct for site-directed mutagenesis and expression.
- Gene Retrieval: Obtain the coding sequence (CDS) of the target gene (e.g., MoLAC14) from the relevant genome or transcriptome database [10].
- Vector Preparation: Use a standard expression vector such as pET-28a. Linearize the vector using appropriate restriction enzymes (e.g., Nde I and Xho I) [10].
- Gene Insertion: Integrate the target gene into the linearized vector using Gibson assembly or similar methods to create the wild-type expression plasmid [10].
- Site-Directed Mutagenesis:
- To introduce alanine mutations, use the Splice-Overlapping Extension PCR (SOE-PCR) method [44].
- Design forward and reverse primers containing the desired alanine codon mutation (GCT, GCC, GCA, or GCG).
- Perform a first-round PCR to generate two overlapping DNA fragments that contain the mutation.
- Use these fragments as templates in a second, overlapping PCR to generate the full-length mutated gene.
- Ligation and Transformation: Ligate the mutated gene into the expression vector and transform into a cloning strain of E. coli (e.g., DH5α). Select transformants on antibiotic-containing plates [44].
- Sequence Verification: Isolate plasmid DNA from positive clones and verify the sequence of the mutated gene through Sanger sequencing [44].
Protein Expression and Purification
This protocol describes the expression and purification of the recombinant wild-type and mutant enzymes.
- Strain Transformation: Transform the sequenced plasmid into an expression host, such as E. coli BL21(DE3) [10] [44].
- Culture and Induction:
- Grow the transformed cells in a suitable medium at 37°C until the optical density at 600 nm (OD₆₀₀) reaches approximately 0.6-0.8.
- Induce protein expression by adding 1 mM IPTG.
- To ensure proper metalloenzyme folding, simultaneously add Copper (II) ions (Cu²âº) to the culture medium [10].
- Continue incubation overnight at a controlled temperature (e.g., 37°C) [44].
- Cell Harvest and Lysis:
- Harvest the cells by centrifugation.
- Resuspend the cell pellet in an appropriate buffer (e.g., 50 mM Tris-HCl, pH 8.0).
- Lyse the cells using sonication or a similar method.
- Clarify the lysate by centrifugation to obtain the soluble protein fraction [44].
- Protein Purification:
- Precipitate the crude protein from the supernatant using ammonium sulfate (e.g., 60% saturation).
- Dialyze the resuspended pellet to remove salts.
- Further purify the protein using ion-exchange chromatography (e.g., a DEAE-Sepharose column with a 0-0.5 M NaCl gradient) or affinity chromatography tailored to the protein's tag [44].
In Vitro Enzyme Activity Assay
This procedure is used to test the ability of the purified laccase to convert chavicol into magnolol.
- Reaction Setup: Prepare a 200 μL reaction mixture containing:
- Reaction Termination: Stop the reaction by adding acetonitrile containing an internal standard [44] [45].
- Sample Analysis:
- Centrifuge the terminated reaction to remove precipitates.
- Analyze the supernatant using HPLC and MS to detect and quantify the magnolol product [10].
- Confirm magnolol identity by comparing its retention time and mass spectrum with an authentic standard.
Data Analysis and Validation
- Quantification: Calculate magnolol concentration based on HPLC peak area calibrated against a standard curve.
- Catalytic Efficiency: Determine kinetic parameters (Kₘ, Vₘâ‚â‚“, kcat) by performing the activity assay with varying substrate concentrations. The catalytic efficiency is given by kcat/Kₘ.
- Thermal Stability: Compare the half-lives (tâ‚/â‚‚) and thermal unfolding curves of wild-type and mutant enzymes using techniques like differential scanning calorimetry to assess the impact of mutations on stability [44].
Workflow and Pathway Diagrams
Alanine Scanning for Enzyme Optimization
Magnolol Biosynthetic Pathway in Synthetic Biology
Resolving Metabolic Burden and Toxicity in Engineered Hosts
The engineering of microbial cell factories for the production of valuable compounds, such as the plant-derived antimicrobial magnolol, is a central goal of synthetic biology. However, redirecting cellular metabolism towards production pathways places significant metabolic burden on the host organism, often triggering stress responses that reduce growth, decrease genetic stability, and limit final product yields [46] [47]. This metabolic burden represents a major bottleneck in the development of economically viable bioprocesses, as it manifests as hidden constraints on host productivity and robustness [48] [47]. In the context of magnolol production, where synthetic biology offers a promising route to circumvent the lengthy cultivation and low yields associated with plant extraction, understanding and mitigating this burden is paramount for success [5]. This Application Note details the underlying causes of metabolic burden and provides validated experimental protocols to identify, quantify, and resolve these challenges, enabling the development of robust and efficient engineered hosts.
Understanding Metabolic Burden: Mechanisms and Symptoms
Fundamental Triggers of Metabolic Burden
Metabolic burden arises from the redirection of cellular resources—including energy molecules (ATP), reducing equivalents, carbon skeletons, and amino acids—away from host growth and maintenance and towards the expression of heterologous pathways and the synthesis of the target product [47]. Engineering strategies such as (over)expression of heterologous proteins and gene knockouts inevitably disturb the highly regulated native metabolism [46]. Key triggers include:
- Resource Competition: Heterologous pathway expression directly competes with native processes for shared, limited cellular resources. This includes the depletion of intracellular amino acid pools and their corresponding charged tRNAs, which can stall translation [46].
- Energetic Inefficiency: Engineering metabolism for bioproduction consumes building blocks and energy molecules, often triggering inherent inefficiencies that reduce the host's fitness [47].
- Toxicity and Stress: The accumulation of non-native or overproduced intermediates, the physical stress of expressing membrane proteins, and the final product's potential cytotoxicity can activate various stress response mechanisms [46].
Observable Physiological Symptoms
The triggers of metabolic burden lead to a range of observable physiological symptoms that negatively impact bioproduction. The relationships between these triggers, the underlying stress mechanisms they activate, and the resulting symptoms are complex and interconnected, as illustrated below.
Table: Key Stress Mechanisms and Their Triggers
| Stress Mechanism | Primary Trigger(s) | Key Actor(s) | Physiological Consequence |
|---|---|---|---|
| Stringent Response [46] | Depletion of amino acids or charged tRNAs in the ribosomal A-site. | RelA/SpoT enzymes; (p)ppGpp alarmones. | Halts rRNA/tRNA synthesis, redirects transcription to amino acid biosynthesis. |
| Heat Shock Response [46] | Accumulation of misfolded proteins. | Chaperones (DnaK, DnaJ); Proteases (FtsH, ClpXP). | Increased degradation of misfolded proteins and inhibition of global protein synthesis. |
| Envelope Stress | Overexpression of membrane proteins; product toxicity. | σE and Cpx pathways. | Disruption of cell envelope integrity, affecting membrane potential and transport. |
Quantitative Assessment of Metabolic Burden
Accurately quantifying the metabolic burden is the first step towards its resolution. The following table summarizes key metrics that should be monitored.
Table: Metrics for Quantifying Metabolic Burden in Engineered Hosts
| Metric Category | Specific Parameter | Measurement Technique | Interpretation |
|---|---|---|---|
| Growth Kinetics | Maximum specific growth rate (µmax) | Spectrophotometry (OD600) | Direct indicator of cellular fitness and health. |
| Lag phase duration | Growth curve analysis | Prolonged lag suggests adaptation to stress. | |
| Productivity | Final product titer (e.g., mg/L magnolol) | HPLC, MS [5] | Ultimate measure of production efficiency. |
| Product yield (YP/S) | Analytics (HPLC), substrate consumption | Stoichiometric efficiency of conversion. | |
| System-Level Physiology | Metabolic Flux Distribution | 13C-MFA [47] | Identifies flux rerouting and bottlenecks. |
| RNA-to-Protein Ratio | RNA quantification, protein assays | Indicator of translational capacity and stress. | |
| Genetic Stability | Plasmid Retention Rate | Plate counting, flow cytometry | Measures loss of engineered traits over generations. |
Protocol 1: Profiling Growth and Production Dynamics
Objective: To simultaneously monitor the impact of metabolic burden on host growth and product formation in a high-throughput manner.
Materials:
- Strains: Control (empty vector) and engineered production strain (e.g., E. coli expressing magnolol biosynthesis genes [5]).
- Equipment: Microplate reader capable of measuring OD and fluorescence/absorbance; deep-well plates; shaking incubator.
- Media: Appropriate defined or complex medium (e.g., LB, M9).
Procedure:
- Inoculum Preparation: Grow overnight cultures of both control and engineered strains in a non-selective medium to avoid confounding stress.
- Dilution and Loading: Dilute overnight cultures to a standard OD600 of 0.05 in fresh, selective medium. Transfer 200 µL aliquots into a 96-well deep-well plate. Include at least 6 biological replicates per strain.
- Growth Curve Monitoring: Place the plate in a pre-warmed (e.g., 37°C) microplate reader with continuous shaking. Measure the OD600 every 15-30 minutes for 24-48 hours.
- Sampling for Product Analysis: At defined time points (e.g., mid-exponential, early stationary, and late stationary phase), remove 100 µL of culture from designated wells.
- Centrifuge samples at high speed to separate cells from supernatant.
- Analyze the supernatant or cell lysate (depending on product localization) for magnolol production via HPLC or the applicable method [5].
- Data Analysis:
- Calculate µmax from the linear region of the ln(OD600) versus time plot.
- Plot product titer against time and OD to understand production phase (growth-associated vs. non-growth associated).
Strategies to Relieve Metabolic Burden
A multi-faceted approach is required to mitigate metabolic burden. The strategies below can be implemented individually or in combination.
Table: Summary of Metabolic Burden Mitigation Strategies
| Strategy | Core Principle | Example Application | Key Benefit |
|---|---|---|---|
| Dynamic Pathway Regulation [48] [47] | Decouples cell growth from production phase using quorum-sensing, metabolite-sensing, or inducible promoters. | Inducing magnolol production (e.g., via laccase expression [5]) only after high cell density is achieved. | Maximizes biomass accumulation before diverting resources to production. |
| Genomic Integration [47] | Replaces high-copy plasmids by stably integrating genes into the host chromosome. | Using CRISPR-Cas [49] to integrate key magnolol biosynthetic genes (e.g., MoLAC14 [5]) into the genome. | Eliminates plasmid-related burden (replication, selection) and improves genetic stability. |
| Enzyme & Pathway Optimization | Optimizes codon usage [46], uses weaker promoters, or engineers enzymes for higher stability/activity. | Engineering MoLAC14 laccase with E345P/G377P mutations for enhanced thermal stability [5]. | Reduces the resource cost per functional enzyme molecule, improving efficiency. |
| Enhancing Energy Metabolism [47] | Increases ATP and reducing equivalent supply by overexpressing respiratory chain components. | Modifying central carbon metabolism to increase NADPH availability for P450 enzymes in precursor pathways. | Alleviates energy bottlenecks that constrain both growth and production. |
| Consortium Engineering [48] | Divides a complex metabolic pathway across multiple, specialized microbial strains. | One strain produces the magnolol precursor chavicol, while a second, optimized strain performs the laccase-mediated coupling to magnolol [5]. | Distributes the burden, preventing any single host from being overloaded. |
Protocol 2: Implementing a Quorum-Sensing Mediated Dynamic Control System
Objective: To decouple cell growth from product formation by placing a key metabolic bottleneck gene under the control of a quorum-sensing promoter.
Materials:
- Plasmids: pQS-GFP (Reporter: constitutively expressed RFP; PluxI-GFP). pPROD-MoLAC14 (Production: constitutively expressed LuxI; PluxI-MoLAC14) [5].
- Strains: E. coli production host.
- Reagents: AHL (N-(3-Oxohexanoyl)-L-homoserine lactone), antibiotics.
Procedure:
- Circuit Assembly: Clone the gene of interest (e.g., the magnolol-synthesizing laccase MoLAC14 [5]) downstream of a AHL-inducible Plux promoter in a suitable vector to create pPROD-MoLAC14.
- Strain Transformation: Co-transform the production host with the pPROD-MoLAC14 plasmid and a reporter plasmid (pQS-GFP) to monitor circuit activation.
- Characterization in Bioreactor:
- Inoculate a fermenter with the engineered strain and monitor OD600 and GFP fluorescence.
- As the culture grows, it endogenously produces the AHL signal (via LuxI). Once a high cell density (e.g., OD600 > 5) and a corresponding threshold of GFP are reached, sample the culture to confirm MoLAC14 expression and magnolol production via SDS-PAGE and HPLC, respectively [5].
- Comparison: Run a control fermentation with a strain where MoLAC14 is constitutively expressed. Compare the growth curves and final magnolol titers between the dynamic and constitutive systems.
The logical workflow for designing a burden-relieved strain, from initial design to final validation, is summarized in the following diagram.
Case Study: Applying Burden Mitigation to Magnolol Production
The production of magnolol in a microbial host like E. coli presents an ideal scenario for applying these principles. The hypothesized pathway involves the laccase-mediated coupling of chavicol, which is derived from tyrosine [5].
The Challenge: Constitutively expressing the entire pathway, especially the laccase MoLAC14, is likely to impose significant burden through resource competition and potential toxicity of intermediates or the final product.
Proposed Solution:
- Divide Pathway: Split the pathway. Engineer a first strain to overproduce the precursor chavicol from tyrosine. Engineer a second strain specializing in the conversion of chavicol to magnolol via an optimized MoLAC14 laccase.
- Optimize the Key Enzyme: Use the engineered MoLAC14 variant with mutations like L532A, which was shown to boost magnolol production to 148.83 mg/L [5], thereby reducing the required expression level for a given titer.
- Implement Dynamic Control: In the magnolol-producing strain, place the optimized MoLAC14 gene under a quorum-sensing promoter. This ensures expression is triggered only at high cell density, preventing unnecessary burden during the growth phase.
The Scientist's Toolkit: Key Reagents for Magnolol Bioproduction
Table: Essential Research Reagents for Engineering Magnolol Production
| Reagent / Tool | Function / Purpose | Example from Literature |
|---|---|---|
| Laccase Gene (MoLAC14) [5] | Catalyzes the oxidative coupling of two chavicol molecules to form magnolol. | MoLAC14 from Magnolia officinalis; engineered variants (e.g., L532A) for higher activity. |
| pET-28a Vector [5] | A common protein expression vector for cloning and high-level expression in E. coli. | Used for heterologous expression and functional analysis of candidate laccase genes. |
| Chavicol | The direct precursor molecule for magnolol synthesis. | Used as a substrate in in vitro enzymatic assays to validate laccase activity [5]. |
| HPLC & Mass Spectrometry [5] [7] | Analytical techniques for identifying and quantifying magnolol production in culture supernatants or lysates. | Critical for validating successful biosynthesis and measuring titers. |
| Quorum-Sensing Plasmid System | Enables dynamic, population-density-dependent gene expression. | A Plux-based system to control laccase expression and decouple it from growth. |
Metabolic burden is an inevitable challenge in metabolic engineering, but it can be systematically managed. By quantitatively profiling host physiology and implementing rational strategies such as dynamic regulation, genomic integration, and enzyme engineering, it is possible to design robust microbial cell factories that maintain high viability and productivity. Applying these principles to the nascent field of microbial magnolol production provides a structured framework to overcome inherent bottlenecks, paving the way for the efficient and sustainable synthesis of this and other high-value plant-derived compounds.
The transition from laboratory-scale cultivation in shake flasks to controlled production in industrial bioreactors represents a critical, and often challenging, phase in the development of bioprocesses, particularly within the field of synthetic biology. For the sustainable production of valuable compounds like magnolol—a natural biphenyl with documented antimicrobial, anti-inflammatory, and anticancer properties—overcoming these scalability challenges is paramount [8] [3]. Traditional extraction of magnolol from Magnolia officinalis bark is constrained by the plant's long cultivation time and low compound concentration (approximately 1%), making synthetic biology an attractive alternative [10]. However, the path from a genetically engineered strain in a shake flask to a robust industrial process is fraught with technical obstacles related to environmental control, mixing, and mass transfer. This application note details these scalability challenges within the context of magnolol production, providing a structured comparison of cultivation systems, detailed experimental protocols for process characterization, and a strategic framework for successful scale-up.
Key Differences Between Shake Flasks and Bioreactor Systems
Understanding the fundamental operational differences between shake flasks and bioreactors is the first step in de-risking scale-up. The table below summarizes the critical parameters that diverge significantly between these two systems.
Table 1: System Comparison for Microbial Cultivation
| Parameter | Shake Flasks | Bioreactors |
|---|---|---|
| Primary Function | Screening, initial strain and media development [50] | Process optimization, scale-up, and production [50] |
| Process Control | Limited and indirect; via incubator environment [51] | Direct, real-time monitoring and control of multiple parameters [50] [51] |
| pH Control | Not available; buffered media only [52] | Precise, automated control via acid/base addition [51] |
| Dissolved Oxygen (DO) | Uncontrolled; depends on flask geometry and shaking speed [50] | Precisely controlled via stirrer speed, gas blending, and air flow [50] [51] |
| Mixing Mechanism | Orbital shaking [50] | Mechanical stirring with impellers [50] |
| Feeding Strategies | Limited to batch or manual addition [51] | Automated fed-batch or continuous perfusion [51] |
| Maximum Cell Density (E. coli OD600) | ~4-6 (Batch) [51] | ~40 (1-day Fed-Batch) to ~230 (2-day Fed-Batch) [51] |
| Scale-Up Basis | Not directly scalable [53] | Based on engineering parameters (e.g., P/V, kLa) [53] |
The consequences of these differences are profound. The inability to control pH and dissolved oxygen in shake flasks means that cells experience a dynamic, non-optimal environment, which can alter their physiology and metabolic output [52]. In contrast, the precise control afforded by bioreactors supports higher cell densities and more consistent productivity, as evidenced by the order-of-magnitude higher cell densities achievable with E. coli [51]. For a process aimed at producing a target molecule like magnolol, this translates directly to higher titers and a more predictable process.
Experimental Protocols for Scalability Assessment
Protocol: Determining the Oxygen Transfer Coefficient (kLa)
Objective: To quantify the oxygen mass transfer capacity of both shake flasks and a bench-scale bioreactor, providing a key scale-up criterion [53].
Principle: The kLa (volumetric oxygen transfer coefficient) defines the maximum oxygen transfer rate into the culture. It is a critical parameter for ensuring aerobic conditions are maintained at increasing scales.
Materials:
- Dissolved oxygen (DO) probe (calibrated to 0% and 100% air saturation)
- Bench-top bioreactor system
- Shake flasks with adapters for DO probes
- Sodium sulfite (Na₂SO₃) solution (0.5 M) in a suitable buffer (e.g., 0.1 M phosphate buffer)
- Cobalt chloride (CoClâ‚‚) catalyst solution (0.001 M)
Method:
- Dynamic Gassing-Out Method (for Bioreactors): a. Fill the bioreactor with a model fluid (e.g., water or culture media). b. Sparge the liquid with nitrogen gas to deplete dissolved oxygen to 0%. c. Once stable at 0%, switch the gas supply to air and start the agitator. d. Record the increase in DO percentage over time until it stabilizes at 100%. e. Plot ln(1 - DO/100) versus time. The kLa is the negative slope of the resulting linear plot.
- Sulfite Oxidation Method (for Shake Flasks): a. Fill the shake flask with a known volume of sodium sulfite solution with cobalt chloride catalyst. b. Place the flask on the shaker under standard operating conditions (speed, fill volume, temperature). c. The sulfite ions (SO₃²â») are oxidized to sulfate (SO₄²â») by dissolved oxygen, which is a zero-order reaction with respect to oxygen concentration. d. Take samples at regular time intervals and titrate with a standard iodine solution to determine the residual sulfite concentration. e. The rate of sulfite oxidation is equivalent to the oxygen transfer rate (OTR). kLa can be calculated as OTR/C, where C is the saturation concentration of oxygen in the liquid.
Protocol: Assessing Strain Performance and Magnolol Titration
Objective: To evaluate the performance of a magnolol-producing strain under different cultivation conditions and quantify the final product yield.
Principle: This protocol leverages the Design-Build-Test-Learn (DBTL) cycle, a core principle in synthetic biology strain engineering [54]. A strain engineered for magnolol production (e.g., E. coli or P. pastoris expressing the key laccase enzyme MoLAC14) is tested in parallel systems to gather data for scaling [10].
Materials:
- Engineered production strain (e.g., E. coli BL21(DE3) with pET-28a-MoLAC14 plasmid) [10]
- LB or defined production media
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) for induction
- Chavicol (enzymatic substrate for MoLAC14) [10]
- Copper (II) ions (Cu²âº, cofactor for laccase activity) [10]
- High-Performance Liquid Chromatography (HPLC) system with Mass Spectrometry (MS) detection
Method:
- Inoculum Preparation: Inoculate a single colony of the production strain into shake flasks containing media and grow overnight to a standard optical density (e.g., OD600 ~2).
- Production Cultivation: a. Shake Flask: Transfer the inoculum to production flasks. Induce with IPTG and add chavicol and Cu²⺠at the appropriate time. b. Bioreactor: Transfer the inoculum to the bioreactor. Maintain precise control over temperature, pH, and DO. Induce and add substrates as per the shake flask protocol.
- Process Monitoring: Sample the culture periodically to measure OD600 (cell density), substrate consumption (e.g., chavicol via HPLC), and potential by-products.
- Product Quantification: a. Centrifuge culture samples to remove cells. b. Extract magnolol from the supernatant using an organic solvent (e.g., ethyl acetate). c. Analyze the extract using HPLC-MS. Quantify magnolol by comparing the peak area to a standard curve generated with authentic magnolol standard. d. Report the final titer in mg/L. The benchmark from recent research is 148.83 mg/L using an engineered MoLAC14 enzyme [10].
Table 2: Research Reagent Solutions for Magnolol Bioproduction
| Reagent / Material | Function in the Experiment |
|---|---|
| Chavicol | The direct precursor substrate for the laccase (MoLAC14) in the one-step synthesis of magnolol [10]. |
| pET-28a Vector | An E. coli expression plasmid used to clone and express the MoLAC14 gene under an inducible promoter [10]. |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | A chemical inducer that triggers the expression of the MoLAC14 gene in the pET system [10]. |
| Copper (II) Ions (Cu²âº) | An essential cofactor required for the catalytic activity of the laccase enzyme MoLAC14 [10]. |
| Laccase Enzyme MoLAC14 | The key biocatalyst identified in Magnolia officinalis that dimerizes chavicol to form magnolol [10]. |
Scale-Up Strategies and Implementation
Scaling a bioprocess is not a simple matter of increasing volume. The objective is to maintain a constant physiological environment for the cells to ensure consistent productivity and product quality. The following diagram illustrates the core logical workflow for navigating the scale-up journey.
Scale-Up Strategy Workflow
The choice of scale-up criterion involves critical trade-offs, as maintaining one parameter constant will cause others to change non-linearly [53]. The table below outlines these trade-offs for common strategies when scaling up from a small bench-scale reactor to a larger pilot or production reactor.
Table 3: Analysis of Common Bioreactor Scale-Up Criteria and Trade-Offs
| Scale-Up Criterion | Impact on Other Parameters | Suitability for Magnolol Production |
|---|---|---|
| Constant Power per Unit Volume (P/V) | Increases impeller tip speed and shear; increases kLa; increases mixing time [53]. | Good for shear-tolerant microbes (e.g., E. coli, P. pastoris); helps maintain oxygen levels for high-density cultures. |
| Constant Oxygen Transfer Coefficient (kLa) | Decreases power input per unit volume; can lead to poorer mixing and gradients [53]. | Critical if magnolol production is highly oxygen-sensitive. Must be checked for mixing limitations. |
| Constant Impeller Tip Speed | Significantly reduces P/V; can drastically lower kLa and increase mixing time [53]. | Used to control shear stress, often for sensitive mammalian cells. Less common for microbial magnolol production. |
| Constant Mixing Time | Requires a massive, often infeasible, increase in P/V [53]. | Not practical as a primary criterion, but mixing time should be evaluated to avoid substrate gradients. |
For a magnolol production process, a hybrid approach is often most effective. The initial scale-up may target a constant kLa to ensure the aerobic laccase enzyme [10] has sufficient oxygen, while simultaneously monitoring metrics like productivity and yield to ensure the scale-dependent parameters remain within an acceptable window.
Navigating the path from laboratory shakes to industrial bioreactors is a complex but manageable engineering endeavor. For the synthetic biological production of magnolol, success hinges on recognizing the profound limitations of shake flasks and systematically employing engineering principles to recreate and control the optimal production environment at larger scales. By quantitatively characterizing processes using parameters like kLa, strategically selecting scale-up criteria based on the organism's biology and process needs, and leveraging the precise control afforded by bioreactors, researchers can overcome scalability challenges. This enables the transition from a promising proof-of-concept in a shake flask to a robust, economically viable industrial bioprocess capable of meeting the demand for magnolol and other high-value natural products.
Benchmarking Success: Analytical Validation and Comparative Analysis of Production Platforms
Within the rapidly advancing field of synthetic biology for magnolol production, robust analytical methods are paramount for validating the output and quality of bioengineered systems. The transition from traditional plant extraction to microbial or enzymatic production of magnolol, a bioactive neolignan from Magnolia officinalis, necessitates reliable techniques for quantifying target compounds and identifying impurities [10] [5]. This document details standardized protocols for High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS) methods, providing a critical analytical framework for researchers developing synthetic biology platforms for magnolol and its isomers.
Core Analytical Techniques and Their Applications
The quantification and identification of magnolol and its regioisomer honokiol are primarily achieved through chromatographic and spectrometric methods. The table below summarizes the key parameters for the established techniques.
Table 1: Summary of Analytical Methods for Magnolol and Honokiol Detection
| Method | Separation Conditions | Detection & Quantitation | Key Performance Metrics | Primary Application |
|---|---|---|---|---|
| TLC-Densitometry [55] [56] | Stationary Phase: Silica gel platesMobile Phase: n-hexane - ethyl acetate - ethanol (16:3:1, v/v/v) | Detection: UV absorbance, λ = 290 nm | Magnolol: LOD 90 ng/zone, LOQ 280 ng/zoneHonokiol: LOD 70 ng/zone, LOQ 200 ng/zone | Quality control of dietary supplements; stability assessment. |
| HPLC-UV [57] | Column: C18 (150 mm × 4.6 mm, 5 μm)Mobile Phase: Methanol:Water (60:40, v/v)Flow Rate: 0.8 mL/min | Detection: UV, λ = 230 nm | Excellent linearity (R² > 0.999), high precision (%RSD < 1%) | Stability-indicating method for bulk substance and formulations. |
| LC-MS/MS [58] | Column: Reversed-phase C18Mobile Phase: Acetonitrile:Water (75:25, v/v)Flow Rate: 0.8 mL/min | Detection: Tandem MS, SRM modeIon Transitions:Honokiol: m/z 265 → 224Magnolol: m/z 265 → 247 | Linear range: 0.0025-0.5 μg/mL (R² > 0.995); high sensitivity and specificity | Sensitive quantification in plant extracts and biological matrices. |
| HPTLC-EDA-MS [55] [56] | HPTLC plates with same mobile phase as TLC | Detection: Bioautography (Antioxidant, Antibacterial), followed by ESI-MS | Confirms bioactivity and identity simultaneously; identifies other bioactive compounds (e.g., piperine). | Bioactivity profiling of complex mixtures like dietary supplements. |
Detailed Experimental Protocols
Protocol 1: Quantitative Analysis of Magnolol and Honokiol by LC-MS/MS
This method provides high sensitivity and specificity for the simultaneous determination of magnolol and honokiol, ideal for validating production yields in synthetic biology approaches [58].
I. Materials and Reagents
- Analytical Standards: Magnolol and honokiol (purity ≥ 95%).
- Solvents: LC-MS grade acetonitrile, water, and methanol.
- Mobile Phase: Acetonitrile and water (75:25, v/v).
- Equipment: HPLC system coupled with a tandem mass spectrometer equipped with an electrospray ionization (ESI) source.
II. Sample Preparation
- Standard Solutions: Accurately weigh 1 mg of each magnolol and honokiol standard. Dissolve in methanol and dilute to a final concentration of 1 μg/mL.
- Test Samples:
- For Magnolia bark extracts: Dilute the extract in methanol.
- For microbial fermentation broth from synthetic biology: Centrifuge the broth to remove cells. Extract the supernatant with an equal volume of ethyl acetate. Evaporate the organic layer to dryness and reconstitute in methanol for analysis [10].
III. Instrumental Parameters and Analysis
- Chromatography:
- Column: Reversed-phase C18 column.
- Mobile Phase: Isocratic elution with acetonitrile:water (75:25, v/v).
- Flow Rate: 0.8 mL/min.
- Injection Volume: 10 μL.
- Mass Spectrometry:
- Ionization Mode: ESI in positive mode.
- Operation Mode: Selected Reaction Monitoring (SRM).
- SRM Transitions:
- Honokiol: m/z 265.0 → 224.0
- Magnolol: m/z 265.0 → 247.0
- Optimize collision energies and source parameters for maximum signal intensity.
IV. Validation and Quantification
- Calibration Curve: Prepare and analyze a series of standard solutions (e.g., 0.0025, 0.01, 0.05, 0.1, 0.5 μg/mL) in triplicate.
- Plot the peak area against the concentration for each analyte to generate a calibration curve. The method demonstrates excellent linearity (R² > 0.995) over this range [58].
Protocol 2: HPTLC-Effect Directed Analysis (EDA) for Bioactivity Validation
This protocol is essential for correlating chemical identity with biological activity, crucial for confirming the functionality of biosynthetically produced magnolol [55] [56].
I. Materials and Reagents
- HPTLC Plates: Silica gel 60 F254.
- Mobile Phase: n-hexane - ethyl acetate - ethanol (16:3:1, v/v/v).
- Detection Reagents:
- DPPH solution (0.04% in methanol) for antioxidant activity.
- Microbial Suspensions: Bacillus subtilis or Aliivibrio fischeri for antibacterial bioautography.
II. Separation and Bioautography
- Application: Apply test samples and standards as bands on the HPTLC plate.
- Development: Develop the plate in a pre-saturated twin-trough chamber with the mobile phase to a distance of 70 mm.
- Derivatization/Bioassay:
- Antioxidant Assay: Dip the developed plate in the DPPH solution. Active compounds appear as yellow bands against a purple background.
- Antibacterial Assay: Cover the developed plate with a soft agar layer inoculated with the test bacterium. After incubation, clear zones of inhibition indicate antibacterial activity.
III. HPTLC-MS Interface
- After documentation of bioautography, target zones (e.g., magnolol/honokiol) can be eluted directly from the plate using an elution-based interface connected to a mass spectrometer.
- MS Conditions: Use ESI-MS in positive ion mode to confirm the identity of the bioactive compounds based on their molecular ion ([M+H]+ for magnolol/honokiol is m/z 265) and fragmentation pattern [55].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents and Materials for Magnolol Analysis and Biosynthesis
| Item/Category | Function/Application | Specific Examples / Notes |
|---|---|---|
| Laccase Enzymes | Catalyzes the oxidative coupling of chavicol to form magnolol in synthetic pathways. | MoLAC14: A key laccase from M. officinalis identified for magnolol synthesis. Mutants like E345P show enhanced thermal stability [10] [5]. |
| Chromatography Columns | Separation of magnolol from honokiol and other impurities in complex mixtures. | Reversed-Phase C18 Column (e.g., 150 mm x 4.6 mm, 5 μm) is standard for HPLC analysis [58] [57]. |
| Mass Spectrometry Standards | Instrument calibration and creation of quantitative calibration curves. | High-purity magnolol and honokiol analytical standards are essential for accurate LC-MS/MS quantification [58]. |
| Bioassay Reagents | Linking chemical presence to biological function via HPTLC-EDA. | DPPH (2,2-Diphenyl-1-picrylhydrazyl): A stable free radical for antioxidant activity screening on HPTLC plates [55] [56]. |
Workflow and Pathway Visualization
The following diagram illustrates the integrated workflow for the production of magnolol via synthetic biology and its subsequent analytical validation, highlighting the critical role of the methods described in this document.
Integrated Workflow for Magnolol Production and Analysis
The precise and accurate analytical methods detailed herein—particularly HPLC and Mass Spectrometry—are indispensable for the advancement of synthetic biology routes for magnolol production. They provide the necessary tools to quantify titers, confirm structural identity, assess purity and stability, and validate biological activity. By adopting these standardized protocols, researchers can robustly compare and optimize different production strains and processes, accelerating the development of a sustainable and efficient supply chain for this valuable natural compound.
Within synthetic biology, achieving high-titer production is a critical milestone that bridges laboratory-scale research and industrially viable bioprocesses. For valuable natural products like magnolol, a bioactive neolignan from Magnolia officinalis, high-titer production is a prerequisite for making therapeutic applications feasible. Traditional extraction from magnolia bark yields magnolol at concentrations of approximately 1%, presenting significant challenges for supply [59]. This Application Note details a synthetic biology protocol that achieved a groundbreaking magnolol titer of 148.83 mg/L from the precursor chavicol using a engineered laccase enzyme (MoLAC14) in a microbial production system [10]. The strategy centered on the identification and subsequent protein engineering of a key biosynthetic enzyme to enhance its stability and catalytic efficiency, providing a robust framework for the high-yield bioproduction of other complex plant-derived natural products.
Experimental Protocols
Identification of the Key Biosynthetic Enzyme MoLAC14
Principle: This protocol identifies laccase enzymes catalyzing magnolol synthesis from chavicol by analyzing transcriptome data and validating candidate gene function in vitro [10].
Procedure:
- Transcriptome Sequencing & Assembly: Collect diverse tissues (e.g., bark, leaves) from M. officinalis. Perform RNA extraction and transcriptome sequencing (RNA-seq). Assemble the RNA-seq reads and map them to the M. officinalis genome.
- Gene Identification & Annotation: Annotate the assembled transcripts for biological function. Identify all putative laccase genes in the M. officinalis genome.
- Candidate Gene Selection: Analyze gene expression levels (TPM) across tissues. Prioritize laccase genes that are highly expressed in magnolol-producing tissues (e.g., roots, leaves, bark) and those located within potential biosynthetic gene clusters (e.g., MoLAC4, MoLAC17).
- In Vitro Functional Validation: a. Gene Synthesis & Cloning: Synthesize the coding sequences (CDS) of candidate genes (e.g., MoSKU5F, MoLAC7B, MoLAC14, MoLAC4A, MoLAC4B, MoLAC17F). Clone these genes into a protein expression vector (e.g., pET-28a) between Nde I and Xho I restriction sites [10]. b. Recombinant Protein Expression: Chemically transform the constructed plasmids into E. coli BL21(DE3). Culture the expression strains at 37°C and induce protein expression with Isopropyl β-D-1-thiogalactopyranoside (IPTG). Simultaneously add Copper (II) ions (Cu²âº) to the culture medium to facilitate proper laccase folding and metallation. c. Enzyme Activity Assay: Purify the recombinant laccase proteins. Incubate the enzymes with the substrate chavicol in a suitable reaction buffer. Detect and quantify the magnolol product using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS).
Protein Engineering of MoLAC14 for Enhanced Titer
Principle: This protocol enhances magnolol production by improving the thermal stability and activity of the key laccase MoLAC14 through rational design and site-saturation mutagenesis [10].
Procedure:
- Thermal Stability Engineering: a. Perform selective mutations targeting residues within the protein's core and flexible loops to improve rigidity. b. Measure the impact of mutations (e.g., E345P, G377P, H347F, E346C, E346F) on enzyme thermal stability. These specific mutations were shown to notably enhance stability [10].
- Alanine Scanning for Activity Enhancement: a. Systematically mutate solvent-accessible residues to alanine to identify positions critical for catalytic activity or substrate binding. b. Screen the generated mutant libraries for increased magnolol production in a suitable in vivo or in vitro system.
- Validation of High-Producing Mutant: a. Culture the production strain harboring the top-performing mutant (e.g., MoLAC14-L532A) under optimized conditions. b. Quantify the final magnolol titer, which reached 148.83 mg/L for the L532A mutant, using HPLC [10].
Table: Key Mutations in MoLAC14 and Their Effects on Magnolol Production
| Mutation | Primary Effect | Impact on Magnolol Titer |
|---|---|---|
| E345P | Enhanced thermal stability | Increased stability |
| G377P | Enhanced thermal stability | Increased stability |
| H347F | Enhanced thermal stability | Increased stability |
| E346C | Enhanced thermal stability | Increased stability |
| E346F | Enhanced thermal stability | Increased stability |
| L532A | Enhanced enzymatic activity | Production increased to 148.83 mg/L |
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Reagents for Magnolol Biosynthesis Research
| Item | Function/Description | Example/Reference |
|---|---|---|
| pET-28a Vector | Protein expression vector for recombinant enzyme production in E. coli | [10] |
| E. coli BL21(DE3) | Microbial host for recombinant protein expression and bioconversion | [10] |
| Chavicol | Direct precursor substrate for magnolol synthesis | [10] |
| Laccase Enzyme (MoLAC14) | Key oxidase catalyst that dimerizes chavicol to form magnolol | [10] |
| Copper (II) Ions (Cu²âº) | Essential cofactor for laccase enzyme activity and proper folding | [10] |
| High-Performance Liquid Chromatography (HPLC) | Analytical method for quantifying magnolol titer and reaction conversion | [10] |
| Mass Spectrometry (MS) | Method for definitive identification of magnolol structure | [10] |
Workflow and Pathway Diagrams
Diagram: High-Titer Magnolol Production Workflow
Diagram: Engineered Biosynthetic Pathway from Chavicol to Magnolol
Magnolol, a hydroxylated biphenyl compound primarily extracted from the bark of Magnolia officinalis, has garnered significant scientific interest due to its diverse pharmacological properties, including anticancer, antimicrobial, anti-inflammatory, and neuroprotective activities [8] [28]. However, its clinical application and industrial production face challenges related to the compound's poor water solubility, low bioavailability, and the ecological constraints of traditional sourcing methods [8] [28]. This analysis examines the three primary production paradigms—plant extraction, chemical synthesis, and emerging biosynthetic approaches—evaluating their efficiencies, challenges, and potential for sustainable magnolol production to support pharmaceutical and research applications.
Plant Extraction
2.1.1 Principle and Workflow Traditional plant extraction isolates magnolol directly from Magnolia bark using physical and chemical methods. The general workflow involves plant material collection, drying, grinding, extraction using solvents or mechanochemical assistance, and final purification.
2.1.2 Key Protocols
- Mechanochemical Extraction (MCAE): Grind Magnolia bark with solid alkaline reagents (e.g., sodium carbonate at 2.0 wt%) in a ball mill. This mechanical force disrupts plant cell walls and facilitates the conversion of magnolol into a water-soluble salt. The magnolol salt is then extracted with warm water (40°C), followed by acidification to regenerate magnolol [60].
- Ultrasound-Assisted Extraction (UAE): Excite plant material in solvent using ultrasonic waves, enhancing cell wall disruption and mass transfer [8].
- Supercritical Fluid Extraction (SFE): Use supercritical COâ‚‚ as a non-toxic, tunable solvent for efficient and selective extraction [8].
Table 1: Performance Metrics of Plant Extraction Methods
| Extraction Method | Reported Yield | Key Advantages | Major Limitations |
|---|---|---|---|
| Mechanochemical (MCAE) | ~7.1% increase over heat-reflux [60] | High selectivity, minimal organic solvent, simpler purification [60] | Requires specialized equipment, additional reagent input |
| Ultrasound-Assisted (UAE) | Information Missing | Reduced extraction time, moderate equipment cost [8] | Scalability challenges, solvent consumption |
| Supercritical Fluid (SFE) | Information Missing | Green solvent (COâ‚‚), high purity extracts [8] | High capital cost, process optimization complexity |
| Conventional Heat-Reflux | Baseline | Simple setup, well-established [60] | High energy, high solvent consumption, low selectivity [60] |
Chemical Synthesis
2.2.1 Principle and Workflow Chemical synthesis constructs the magnolol molecule de novo from simpler phenolic precursors through controlled chemical reactions, such as coupling and functional group transformations.
2.2.2 Key Protocols Traditional synthesis often starts with phenol or chavicol, using oxidative coupling strategies. For instance, Clark et al. proposed synthesizing magnolol from phenol, but this route suffered from low yield and poor specificity, generating undesirable by-products [5]. A typical laboratory-scale synthesis involves:
- Substrate Preparation: Dissolve the phenolic starting material (e.g., chavicol) in an appropriate organic solvent.
- Oxidative Coupling: Add a chemical oxidant (e.g., FeCl₃, enzyme-mimetic complexes) to the reaction mixture with stirring under controlled temperature and inert atmosphere to facilitate the biphenyl bond formation.
- Reaction Monitoring & Work-up: Monitor reaction progress by TLC or HPLC. Upon completion, quench the reaction and extract the crude product.
- Purification: Purify the crude mixture via column chromatography or recrystallization to obtain pure magnolol [5].
Table 2: Performance Metrics of Chemical Synthesis Methods
| Synthetic Approach | Reported Yield | Key Advantages | Major Limitations |
|---|---|---|---|
| From Chavicol (Runeberg) | Information Missing | Conceptually straightforward | Low yield, multiple steps [5] |
| From Phenol (Clark et al.) | Low | Avoids agricultural dependence | Non-specific, multiple by-products, high solvent consumption [5] |
Biosynthesis
2.3.1 Principle and Workflow Biosynthesis leverages enzymatic machinery—either within the native plant or engineered microbial hosts—to produce magnolol from biological precursors. Recent research has elucidated a potential pathway from tyrosine to magnolol via chavicol, catalyzed by a key laccase enzyme [5].
2.3.2 Key Protocols
- In Vitro Enzymatic Synthesis:
- Gene Identification & Cloning: Identify candidate laccase genes (e.g., MoLAC14) from Magnolia transcriptome data [5]. Clone the codon-optimized gene into an expression vector (e.g., pET-28a).
- Enzyme Expression & Purification: Transform the vector into a host like E. coli, induce protein expression, and purify the recombinant enzyme using affinity chromatography.
- Biotransformation: Incubate the purified MoLAC14 enzyme with the substrate chavicol in a suitable buffer system (e.g., phosphate buffer, pH ~7.0) with mild shaking.
- Product Extraction & Analysis: Extract the reaction mixture with ethyl acetate and concentrate the organic layer. Analyze and quantify magnolol formation using HPLC and Mass Spectrometry [5].
- Enzyme Engineering for Improved Production:
- Site-Directed Mutagenesis: Introduce point mutations (e.g., E345P, G377P, L532A) into the MoLAC14 gene to enhance thermal stability and catalytic activity [5].
- Screening: Express mutant variants, purify them, and screen for improved magnolol production in the biotransformation assay. The L532A mutant achieved a titer of 148.83 mg/L in vitro [5].
Table 3: Performance Metrics of Biosynthesis Methods
| Biosynthesis Method | Reported Yield / Titer | Key Advantages | Major Limitations |
|---|---|---|---|
| In Vitro Enzymatic (MoLAC14) | 148.83 mg/L (L532A mutant) [5] | High specificity, mild reaction conditions, sustainable profile | Requires identified genes, expensive cofactors, process scaling challenges |
| Proposed Plant Biosynthesis | ~1% in bark [5] [28] | Naturally occurring | Very low concentration, long cultivation cycles (10-15 years) [5] |
Integrated Pathway and Workflow Visualization
Magnolol Biosynthetic Pathway
Integrated Production Workflow
The Scientist's Toolkit: Key Research Reagents
Table 4: Essential Reagents and Materials for Magnolol Research and Production
| Reagent/Material | Function/Application | Specific Examples / Notes |
|---|---|---|
| Sodium Carbonate (Na₂CO₃) | Alkaline reagent in MCAE; converts magnolol to water-soluble salt for selective extraction [60]. | Used at 2.0 wt% relative to plant material [60]. |
| Chavicol | Key biosynthetic precursor and starting material in chemical synthesis [5]. | Substrate for in vitro biotransformation with MoLAC14 laccase [5]. |
| Recombinant Laccase (MoLAC14) | Key oxidase enzyme catalyzing oxidative coupling of chavicol to form magnolol [5]. | Can be engineered (e.g., L532A mutant) for enhanced stability and yield [5]. |
| pET-28a Vector | Prokaryotic expression vector for cloning and expressing target enzymes in E. coli [5]. | Used for heterologous expression of MoLAC14 [5]. |
| Ethyl Acetate | Organic solvent for extracting magnolol from plant material or aqueous reaction mixtures [60] [7]. | Preferred for its effectiveness in extracting lignans [7]. |
This comparative analysis reveals a clear trajectory in magnolol production technology. While plant extraction, particularly advanced methods like MCAE, currently offers a direct route with improved selectivity, it remains tethered to biological cultivation cycles. Traditional chemical synthesis, though independent of agricultural constraints, is hampered by inefficiency and environmental concerns. Biosynthesis emerges as the most promising frontier, offering unparalleled specificity and a sustainable profile. The identification and engineering of key enzymes like MoLAC14 represent a pivotal step towards microbial factory production. Future research must focus on optimizing the entire biosynthetic pathway in heterologous hosts, further engineering enzymes for higher activity, and scaling up fermentation processes to realize the full potential of synthetic biology for the robust, green, and economical production of magnolol and its valuable derivatives.
Evaluating Purity, Cost, and Environmental Impact Across Production Methods
Magnolol, a bioactive lignan primarily derived from the bark of Magnolia officinalis, has garnered significant scientific and commercial interest due to its diverse pharmacological properties, including anti-inflammatory, antioxidant, anti-cancer, and antimicrobial activities [8] [3]. The growing demand across pharmaceutical, nutraceutical, and cosmetic industries, with a market projected to reach nearly $360 million by 2032, underscores the need for efficient and sustainable production methods [61]. Traditional approaches, including direct plant extraction and chemical synthesis, face challenges related to resource intensity, environmental impact, and variable product quality [10] [8]. This document, framed within the context of advancing synthetic biology for magnolol production, provides a comparative evaluation of current production methodologies, detailing experimental protocols and analytical techniques essential for researchers and drug development professionals.
Comparative Analysis of Magnolol Production Methods
The table below summarizes the key quantitative and qualitative metrics for the primary production methods, highlighting the trade-offs between purity, cost, and environmental impact.
Table 1: Comparison of Magnolol Production Methods
| Production Method | Reported Yield & Purity | Estimated Cost & Environmental Impact | Key Advantages | Key Limitations |
|---|---|---|---|---|
| Plant Extraction [8] [30] | - Yield: ~1-10% composition in bark [3]- Purity: Requires further purification (e.g., MIL-101(Cr) adsorption) [30] | - Cost: High (long cultivation time, low concentration) [10]- Impact: Resource-intensive, generates herbal extraction residues (HERs) [30] | Direct, traditional method | Low yield, lengthy cultivation, seasonal variation, generates waste |
| Chemical Synthesis [10] [8] | - Yield: Low, with by-products [10]- Purity: Not specific, forms isomers and by-products [10] | - Cost: High energy consumption, solvent recycling [10]- Impact: High; uses organic solvents, causes environmental pollution [10] | Independent of agricultural supply | Low specificity, low yield, hazardous waste, complex separation |
| Synthetic Biology (Laccase-Catalyzed) [10] | - Yield: 148.83 mg/L (with engineered MoLAC14-L532A) [10]- Purity: One-step conversion from chavicol, high specificity [10] | - Cost: Potentially lower with optimized microbial systems- Impact: Green process; mild conditions, aqueous system, high specificity [10] | High specificity, mild reaction conditions, renewable feedstocks | Pathway understanding is limited, requires enzyme/producer optimization |
| Green Integrated Route (from HERs) [30] | - Yield: 8 kg magnolol/ton HERs (coproduct: 1 kg honokiol, 7.64 kg β-amyrin) [30]- Purity: Recovered via MIL-101(Cr) adsorption [30] | - Cost: Low-cost feedstock (waste), total output ~¥170,700/ton HERs [30]- Impact: Cleaner process; comprehensive waste reutilization, reduces pollution [30] | Upcycles industrial waste, high economic output | Requires multiple integrated steps |
Experimental Protocols for Key Production Methods
Protocol: Laccase-Catalyzed Synthesis of MagnololIn Vitro
This protocol is based on the identification and engineering of the pivotal laccase MoLAC14 from M. officinalis [10].
3.1.1. Reagents and Equipment
- Gene Construct: pET-28a vector containing the codon-optimized MoLAC14 gene [10].
- Host Strain: BL21(DE3) E. coli competent cells [10].
- Culture Media: Luria-Bertani (LB) broth and agar plates supplemented with 50 µg/mL kanamycin.
- Induction Reagents: Isopropyl β-D-1-thiogallactopyranoside (IPTG) and Copper (II) sulfate (CuSO₄).
- Substrate: Chavicol (purity >95%).
- Analytical Equipment: HPLC system with PDA detector, Mass Spectrometer (MS), spectrophotometer.
3.1.2. Procedure
- Strain Generation: Chemically transform the constructed pET-28a-MoLAC14 plasmid into BL21(DE3) E. coli cells and plate on selective LB agar. Incubate overnight at 37°C [10].
- Protein Expression:
- Inoculate a single colony into 50 mL of LB broth with kanamycin and grow overnight at 37°C with shaking.
- Dilute the culture 1:100 into fresh, pre-warmed selective LB medium.
- Grow at 37°C until OD₆₀₀ reaches 0.6-0.8.
- Induce protein expression by adding IPTG to a final concentration of 0.1-0.5 mM and CuSOâ‚„ to 0.1-0.5 mM.
- Incubate the culture for 16-20 hours at 16-18°C with shaking [10].
- Enzyme Assay and Magnolol Production:
- Harvest cells by centrifugation (e.g., 4,000 x g for 20 min) and resuspend in a suitable reaction buffer (e.g., phosphate buffer, pH 7.0).
- Use whole cells or prepare a cell-free extract via sonication or lysis.
- To the enzyme preparation, add chavicol substrate to a final concentration of 1-2 mM.
- Incubate the reaction mixture at 30°C with shaking for 2-6 hours.
- Product Analysis:
- Terminate the reaction by extracting with an equal volume of ethyl acetate.
- Analyze the organic phase by HPLC-MS. Magnolol can be identified by its retention time and mass signature and quantified against a standard curve of authentic magnolol [10].
3.1.3. Visualization of Biosynthetic Pathway and Workflow The following diagram illustrates the hypothesized biosynthetic pathway from tyrosine to magnolol and the experimental workflow for its validation.
Protocol: Antiparasitic Bioassay for Magnolol Activity
This protocol outlines the procedures for evaluating the anthelmintic efficacy of magnolol, a key application, using Ascaris suum larvae [7].
3.2.1. Reagents and Equipment
- Parasite Material: Embryonated Ascaris suum eggs isolated from fresh adult worms.
- Media: Hanks’ Balanced Salt Solution (HBSS), RPMI-1640 Medium.
- Test Compound: Magnolol (e.g., purified from extract or commercial source), dissolved in DMSO.
- Controls: Ivermectin (positive control), DMSO (vehicle/negative control).
- Equipment: COâ‚‚ incubator, sonicated water bath, Baermann apparatus, 96-well plates, microscope.
3.2.2. Procedure
- Larval Preparation:
- Isolate eggs from the uteri of female A. suum and embryonate them under appropriate conditions.
- Hatch the embryonated eggs by stirring in HBSS supplemented with 1% penicillin-streptomycin and 0.1% Amphotericin B at 37°C under 5% CO₂ for 30 minutes.
- Separate the larvae from unhatched eggs using a 20 µm mesh and a Baermann apparatus submerged in HBSS. Allow larvae to sediment for 24 hours at 37°C under 5% CO₂ [7].
- Bioassay Setup:
- Resuspend the harvested larvae in RPMI-1640 medium.
- Transfer approximately 50 live, motile larvae into each well of a 96-well plate.
- Add the test compound (magnolol in DMSO). The final concentration of DMSO should not exceed 1% (v/v) of the total media volume. Include a negative control (1% DMSO) and a positive control (e.g., Ivermectin) [7].
- Incubation and Analysis:
- Incubate the plate for 24 hours at 37°C under 5% CO₂.
- After incubation, count the number of live larvae (defined as those exhibiting movement within 4 seconds of observation) in each well.
- Calculate the percentage mortality for each treatment using the formula:
Mortality (%) = [1 - (Live larvae_24h / Live larvae_0h)] × 100%[7].
- Mechanism of Action Studies:
- To investigate the mitochondrial ETC inhibition mechanism, perform RNA-seq analysis on model nematodes (e.g., C. elegans) exposed to sublethal doses of magnolol or conduct functional assays to measure changes in mitochondrial membrane potential [7].
3.2.3. Visualization of Antiparasitic Mechanism The following diagram illustrates the hypothesized primary mechanism of action of magnolol as an anthelmintic.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents and Materials for Magnolol Research
| Reagent/Material | Function/Application | Example & Notes |
|---|---|---|
| Chavicol | Biosynthesis Substrate: Direct precursor for the laccase-catalyzed synthesis of magnolol [10]. | Commercial standard; critical for in vitro enzyme activity assays and pathway validation. |
| MIL-101(Cr) | Adsorption Material: Metal-organic framework used for the selective adsorption and purification of magnolol and honokiol from complex mixtures [30]. | Synthesized material with high absorption capacity (up to 255.64 mg/g); key for green separation from herbal residues. |
| pET-28a Vector | Protein Expression: Prokaryotic expression vector for cloning and heterologously expressing laccase genes in E. coli [10]. | Contains a T7 lac promoter and kanamycin resistance; used for functional characterization of enzymes like MoLAC14. |
| Engineered Yeast Strain | Microbial Production: Platform for the sustainable production of magnolol or related terpenes (e.g., β-amyrin) from hydrolyzed plant biomass [30]. | Engineered Saccharomyces cerevisiae; enables production via synthetic biology and fermentation. |
| Ivermectin | Bioassay Control: Standard anthelmintic drug used as a positive control in antiparasitic activity assays [7]. | Confirms functionality of larval mortality assays; benchmark for evaluating magnolol's efficacy. |
The production landscape for magnolol is evolving from traditional, resource-intensive methods toward more sophisticated, efficient, and sustainable strategies. Synthetic biology, exemplified by the engineered laccase MoLAC14, offers a promising route with high specificity and yield under mild conditions [10]. Concurrently, integrated green routes that valorize herbal extraction residues present an economically and environmentally favorable model for circular production [30]. The experimental protocols and analytical tools detailed herein provide a foundation for researchers to further optimize these processes, characterize the compound's multifaceted biological activities, and advance magnolol towards broader therapeutic and commercial applications.
Assessing the Therapeutic Potential of Bio-produced vs. Natural Magnolol
Magnolol, a hydroxylated biphenyl compound primarily derived from the bark of Magnolia officinalis, has garnered significant scientific interest due to its broad pharmacological properties, including anti-cancer, anti-inflammatory, antimicrobial, antioxidant, and cardioprotective effects [62] [8] [3]. However, its clinical application faces substantial challenges, predominantly due to the limitations of traditional extraction methods from natural sources, which include low yield (approximately 1-10% of dry bark weight), lengthy cultivation cycles (10-15 years), and significant environmental impact from solvent-intensive processes [5] [10] [8]. Advances in synthetic biology present a promising alternative through the bio-production of magnolol, potentially offering a more sustainable, scalable, and efficient manufacturing platform. This Application Note provides a comparative assessment of natural and bio-produced magnolol, detailing experimental protocols and mechanistic insights to guide researchers in therapeutic development.
Comparative Analysis: Natural vs. Bio-produced Magnolol
Table 1: Quantitative Comparison of Natural and Bio-produced Magnolol
| Parameter | Natural Magnolol | Bio-produced Magnolol |
|---|---|---|
| Source | Bark of Magnolia officinalis [8] | Engineered microbial systems (e.g., E. coli) [5] [10] |
| Production Method | Solvent extraction (e.g., methanol, ethyl acetate) [7] [8] | Fermentation of engineered microbes [5] |
| Key Production Step | Plant cultivation, harvesting, and extraction [63] | One-step enzymatic conversion from chavicol via laccase [5] [10] |
| Reported Yield | ~1-10% of dry bark weight [3] | 148.83 mg/L (with engineered MoLAC14 L532A mutant) [5] [10] |
| Production Time | 10-15 years (plant growth) [5] [10] | Days (microbial fermentation) |
| Environmental Impact | High solvent consumption, pollution [5] [10] | Potentially greener, microbial-based process |
| Therapeutic Profile | Well-documented multi-target effects [62] [64] [3] | Expected to be identical (same chemical structure) |
The bio-production of magnolol leverages a recently elucidated biosynthetic pathway. Research has confirmed that magnolol can be synthesized in a one-step conversion from its precursor, chavicol, catalyzed by the enzyme laccase [5] [10]. The identification and engineering of the pivotal laccase enzyme, MoLAC14, from Magnolia officinalis represents a critical breakthrough for synthetic biology approaches. Specific mutations, such as E345P and G377P, have been shown to enhance the thermal stability of MoLAC14, while the L532A mutation significantly boosts magnolol production to unprecedented levels [5] [10].
Therapeutic Mechanisms and Signaling Pathways
Magnolol exerts its diverse therapeutic effects through interaction with multiple cellular signaling pathways. The diagrams below summarize the key anti-cancer and cardioprotective mechanisms.
Anti-Cancer Signaling Pathways
Figure 1: Magnolol's multi-targeted action against cancer cells. It inhibits key proliferation and survival pathways, including NF-κB, PI3K/Akt/mTOR, and MAPK signaling, leading to suppressed tumor growth and metastasis [3]. Its derivative CT2-3 also induces cell cycle arrest and apoptosis in rheumatoid arthritis fibroblast-like synoviocytes via modulating the PI3K/AKT pathway [64].
Cardioprotective and Other Therapeutic Actions
Figure 2: Overview of magnolol's cardioprotective and other therapeutic mechanisms. It mitigates heart injury by improving hemodynamics and reducing oxidative stress, while also exhibiting broad anti-inflammatory, antimicrobial, and neuroprotective properties [62] [7] [8].
Experimental Protocols
Protocol 1: In Vitro Biosynthesis of Magnolol Using Recombinant Laccase
This protocol describes the key method for validating the enzymatic activity of identified laccases (e.g., MoLAC14) in converting chavicol to magnolol [5] [10].
Workflow Overview:
Figure 3: Workflow for the in vitro biosynthesis and validation of magnolol production using a recombinant laccase system.
Detailed Procedure:
Gene Synthesis and Vector Construction:
Recombinant Protein Expression:
- Chemically transform the constructed plasmid into an E. coli BL21(DE3) expression strain [10].
- Culture the transformed strain at 37°C in a suitable medium (e.g., LB broth) with appropriate antibiotics.
- Induce gene expression by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) when the culture reaches the mid-log phase (OD600 ~0.6).
- Concurrently, add Copper (II) ions (Cu²âº) to the culture to facilitate proper laccase folding and activity [10].
Enzyme Purification:
- Harvest the cells by centrifugation after a suitable induction period (e.g., 4-16 hours).
- Lyse the cells and purify the recombinant laccase protein using a method appropriate for the chosen expression system (e.g., affinity chromatography for His-tagged proteins).
In Vitro Enzymatic Reaction:
- Set up a reaction mixture containing the purified laccase enzyme and the substrate chavicol in a suitable buffer.
- Incubate the reaction mixture at the enzyme's optimal temperature to allow for the oxidative coupling of two chavicol molecules into magnolol.
Product Confirmation and Quantification:
- Analyze the reaction products using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry (MS).
- Confirm magnolol production by comparing its retention time and mass spectrum with an authentic standard.
- Quantify the yield of magnolol, typically reported in mg/L of reaction mixture [5] [10].
Protocol 2: Assessing Anti-Cancer Efficacy In Vitro
This standard protocol is used to evaluate the therapeutic potential of both natural and bio-produced magnolol against various cancer cell lines [3].
Detailed Procedure:
Cell Culture:
- Maintain relevant cancer cell lines (e.g., liver, colon, lung, breast) in appropriate media supplemented with fetal bovine serum (FBS) and antibiotics at 37°C in a 5% CO2 atmosphere.
Compound Treatment:
- Prepare a stock solution of magnolol in DMSO. Further dilute it in cell culture medium for treatments, ensuring the final DMSO concentration is non-cytotoxic (typically <0.1%).
- Seed cells in multi-well plates and allow them to adhere overnight.
- Treat the cells with a range of magnolol concentrations. Include a negative control (vehicle only, e.g., 0.1% DMSO).
Viability and Proliferation Assay:
- After an incubation period (e.g., 24, 48, or 72 hours), assess cell viability using standard assays such as the MTT or CCK-8 assay.
- Measure the absorbance according to the kit's protocol and calculate the percentage of cell viability relative to the control. The half-maximal inhibitory concentration (IC50) can be determined from dose-response curves.
Mechanistic Studies:
- Apoptosis Analysis: Use flow cytometry with Annexin V/propidium iodide (PI) staining to quantify apoptotic cells after magnolol treatment.
- Cell Cycle Analysis: Fix and stain cells with PI followed by flow cytometry to determine the distribution of cells in different cell cycle phases (G0/G1, S, G2/M).
- Western Blotting: Analyze protein extraction from treated and control cells to detect changes in the expression and phosphorylation of key signaling molecules (e.g., components of NF-κB, PI3K/Akt, STAT3, and apoptosis-related proteins like PARP and caspases) [3].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Magnolol Research and Development
| Reagent / Material | Function / Application | Specific Example / Note |
|---|---|---|
| Chavicol | Precursor substrate for the enzymatic biosynthesis of magnolol [5] [10] | Critical for in vitro validation of laccase activity and bio-production. |
| pET-28a Vector | Prokaryotic expression vector for recombinant laccase production [5] [10] | Standard system for high-yield protein expression in E. coli. |
| E. coli BL21(DE3) | Host strain for recombinant laccase expression [10] | Suitable for T7 promoter-based protein expression. |
| Cu²⺠Ions | Cofactor supplement for laccase enzyme activity and stability [10] | Added during protein induction to ensure proper metalloenzyme function. |
| HPLC-MS System | Analytical platform for magnolol identification and quantification [5] [7] | Confirms chemical identity and purity of both natural and bio-produced magnolol. |
| Magnolol Standard | Reference compound for analytical calibration and bioactivity comparison [63] | Essential for ensuring accurate quantification and functional equivalence. |
| Cell-Based Assay Kits | Assessment of therapeutic efficacy (e.g., MTT, CCK-8, Annexin V) [3] | Standardized tools for evaluating anti-cancer and other biological activities. |
Synthetic biology-driven bio-production of magnolol presents a formidable and sustainable alternative to traditional plant extraction, effectively addressing critical limitations of yield, scalability, and environmental impact. The identification of the key biosynthetic enzyme laccase (MoLAC14) and its successful engineering for enhanced performance marks a pivotal advancement in the field. The documented therapeutic profile of natural magnolol, with its multi-target mechanisms against cancer, cardiovascular, and autoimmune diseases, provides a strong rationale for the continued development of its bio-produced counterpart. The experimental protocols and research tools detailed in this document offer a foundation for scientists to further explore the equivalence, and potentially superior characteristics, of bio-produced magnolol, accelerating its path into therapeutic applications.
Conclusion
Synthetic biology has fundamentally shifted the paradigm for magnolol production, moving from reliance on slow plant cultivation to precision engineering of microbial cell factories. The identification of the key biosynthetic enzyme MoLAC14 and its subsequent optimization through protein engineering represent foundational breakthroughs, enabling unprecedented production titers. This engineered biosynthetic approach offers a compelling alternative, characterized by superior scalability, sustainability, and potential for cost-effectiveness compared to traditional methods. Future research must focus on comprehensive pathway elucidation, advanced host engineering to maximize yield, and exploring the synthesis of novel, high-value magnolol derivatives with enhanced bioavailability and targeted therapeutic profiles. The successful application of synthetic biology for magnolol paves the way for its use in producing other complex plant-derived natural products, heralding a new era in sustainable drug discovery and development.
References
- 1. Magnolol and its semi-synthetic derivatives [https://pubmed.ncbi.nlm.nih.gov/40335865/]
- 2. Potential role of magnolol: A comprehensive review on its ... [https://www.sciencedirect.com/science/article/pii/S2666459325000514]
- 3. Magnolol and its semi-synthetic derivatives [https://link.springer.com/article/10.1007/s12672-025-02409-2]
- 4. Mitochondrion-targeted Magnolol Derivatives Exert ... [https://www.sciencedirect.com/science/article/abs/pii/S022352342501030X]
- 5. Identification and Validation of Magnolol Biosynthesis ... [https://www.mdpi.com/1420-3049/29/3/587]
- 6. Magnolol as a Radiotherapy Enhancer in Oral Squamous Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12364617/]
- 7. Magnolol and Honokiol Are Novel Antiparasitic Compounds ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12509032/]
- 8. Magnolol: Chemistry and biology [https://www.sciencedirect.com/science/article/abs/pii/S092666902301258X]
- 9. Magnolol and honokiol: potential lead compounds for the ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1578971/full]
- 10. Identification and Validation of Magnolol Biosynthesis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10856379/]
- 11. Identification and Validation of Magnolol Biosynthesis ... [https://pubmed.ncbi.nlm.nih.gov/38338333/]
- 12. Total synthesis, biological evaluation and biosynthetic re ... [https://pubs.rsc.org/sv/content/articlehtml/2024/sc/d4sc03232b]
- 13. Recent Advances in Laccase Production - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC12603382/]
- 14. Laccase-mediated synthesis of bioactive natural products and ... [https://pubs.rsc.org/en/content/articlehtml/2022/cb/d1cb00259g]
- 15. Laccases as green and versatile biocatalysts: from lab to ... [https://bioresourcesbioprocessing.springeropen.com/articles/10.1186/s40643-021-00484-1]
- 16. Engineering the Bacterial Laccase CotA for Functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561958/]
- 17. MiDNE a tool for Multi-omics genes and drugs interactions ... [https://pubmed.ncbi.nlm.nih.gov/41209342/]
- 18. Engineering Escherichia coli and Yeast Chassis for ... [https://www.frontiersin.org/research-topics/75320/engineering-escherichia-coli-and-yeast-chassis-for-synthetic-biology-driven-biosynthesis-and-biobreeding]
- 19. An In-Depth Comparison of Best Protein Expression ... [https://www.alpha-lifetech.com/blog/comparison-of-protein-expression-techniques-for-yield-and-purity/]
- 20. Compare microbial systems for protein expression [https://cultiply.net/en/comparing-microbial-expression-systems/]
- 21. Engineering a Robust Escherichia coli W Platform for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12508785/]
- 22. Construction of a synthetic biology toolkit for the genetic ... [https://www.sciencedirect.com/science/article/pii/S2405805X25000754]
- 23. Construction and Diversification of Natural Product ... [https://pubmed.ncbi.nlm.nih.gov/41070399/]
- 24. Development of a versatile chassis for the efficient ... [https://www.nature.com/articles/s41467-025-62659-0]
- 25. Strategies to Optimize Protein Expression in E. coli - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7162232/]
- 26. Heterologous expression and optimization of the antimicrobial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12535540/]
- 27. Optimization of heterologous protein production process ... [https://www.sciencedirect.com/science/article/abs/pii/S1389172325002208]
- 28. Magnolol and its semi-synthetic derivatives - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12058641/]
- 29. An AI-driven workflow for the accelerated optimization of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12529354/]
- 30. Coproduction of magnolol, honokiol, and β-amyrin from ... [https://www.sciencedirect.com/science/article/pii/S2468025723000249]
- 31. The Future of Metabolic Engineering and Synthetic Biology [https://pmc.ncbi.nlm.nih.gov/articles/PMC3615475/]
- 32. Construction of a synthetic biology toolkit for the genetic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12173524/]
- 33. Magnolol Inhibits the Growth of Cryptococcus neoformans by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12515567/]
- 34. Strategies for discovery and improvement of enzyme function [https://pmc.ncbi.nlm.nih.gov/articles/PMC3815269/]
- 35. Enhancing stability of enzymes for industrial applications [https://pubmed.ncbi.nlm.nih.gov/41060484/]
- 36. Synthetic Biology, Directed Evolution, and the Rational ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10401280/]
- 37. Review Strategies for obtaining stable enzymes [https://www.sciencedirect.com/science/article/pii/003295929385026C]
- 38. Enhancing functional proteins through multimodal inverse ... [https://www.nature.com/articles/s41467-025-65175-3]
- 39. Directed evolution of hydrocarbon-producing enzymes [https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-025-02689-4]
- 40. Restriction Enzyme Tips [https://www.neb.com/en-us/tools-and-resources/usage-guidelines/restriction-enzyme-tips?srsltid=AfmBOoqrJLFD3cQeRtsvGhcIMk0cKrurppEleuJjpdqb_JzKKGZoZkO9]
- 41. Magnolol additive improves growth performance of Linwu ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0259896]
- 42. Alanine scanning - Wikipedia [https://en.wikipedia.org/wiki/Alanine_scanning]
- 43. Alanine Scanning | MyBioSource Learning Center [https://www.mybiosource.com/learn/testing-procedures/alanine-scanning/]
- 44. A Comprehensive Alanine-Scanning Mutagenesis Study Reveals Roles for Salt Bridges in the Structure and Activity of Pseudomonas aeruginosa Elastase - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4376888/]
- 45. Sulfation and Its Effect on the Bioactivity of Magnolol ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9505486/]
- 46. “Metabolic burden†explained: stress symptoms and its related ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10981312/]
- 47. Metabolic Burden: Cornerstones in Synthetic Biology and ... [https://pubmed.ncbi.nlm.nih.gov/26996613/]
- 48. Relieving metabolic burden to improve robustness and ... [https://www.sciencedirect.com/science/article/pii/S0734975024000958]
- 49. Synthetic biology and metabolic engineering paving ... [https://pubs.rsc.org/en/content/articlelanding/2025/ya/d5ya00118h]
- 50. Shaker Flasks vs Bioreactors [https://gmpplastic.com/blogs/useful-articles-on-lab-supplies-faq-section/shaker-flasks-vs-bioreactors?srsltid=AfmBOoqJW6HG_ZvcmMb0ukOtq__gGjIA5lSZi96zt5h9cUx6KU8kzPRU]
- 51. 5 reasons to move from shake flasks to a bioreactor [https://infors-ht.com/en/blog/5-reasons-to-move-from-shake-flasks-to-a-bioreactor]
- 52. Talking about Shake Flasks and Bioreactors [https://www.eppendorf.com/us-en/lab-academy/life-science/cell-biology/it-s-not-just-about-size-talking-about-shake-flasks-and-bioreactors/]
- 53. Exploring Principles of Bioreactor Scale-Up [https://www.bioprocessintl.com/bioreactors/lessons-in-bioreactor-scale-up-part-1-mdash-exploring-introductory-principles]
- 54. Optimizing the strain engineering process for industrial-scale ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10548853/]
- 55. Thin-layer chromatographic quantification of magnolol and ... [https://www.sciencedirect.com/science/article/pii/S0021967320305082]
- 56. Thin-layer chromatographic quantification of magnolol and ... [https://pubmed.ncbi.nlm.nih.gov/32709311/]
- 57. Design and validation of a robust stability-indicating reversed ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12619242/]
- 58. Simultaneous determination of honokiol and magnolol in ... [https://pubmed.ncbi.nlm.nih.gov/16583454/]
- 59. Honokiol and magnolol: A review of structure-activity ... [https://www.sciencedirect.com/science/article/abs/pii/S0031942224001699]
- 60. Efficient and selective extraction of magnolol from ... [https://www.sciencedirect.com/science/article/abs/pii/S0255270111000195]
- 61. Magnolol Market Report | Global Forecast From 2025 To ... [https://dataintelo.com/report/magnolol-market]
- 62. The Cardioprotective Effect of Magnolia officinalis and Its ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12072210/]
- 63. Integrative analyses of the transcriptome and metabolome ... [https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-024-05933-5]
- 64. Magnolol and honokiol: potential lead compounds for the new ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12055524/]